fractionmissing file
25 views
Skip to first unread message
Giacomo Mutti
Sep 21, 2021, 7:26:34 AM9/21/21
to GeneRax
Dear Benoit, first of all thanks for this amazing tool!
I'm sorry for the trivial question but I can't figure out what the fractionmissing.txt file produced by generax is supposed to be.
I expected that values on fraction missing + values on perspeciescoverage sum to 1 for each species but that is not the case (with my data). What does the value in this file represent?
Thanks and have a nice day,
Giacomo Mutti
Benoit Morel
Sep 22, 2021, 2:59:50 AM9/22/21
to GeneRax
Dear Giacomo Mutti,
It should be the proportion of families that contain (or do not contain, I don't remember) at least one gene from the species.
So it should not sum to one.
Does it make sense with your data?
Best,
Benoit
Giacomo Mutti
Sep 22, 2021, 4:00:23 AM9/22/21
to Benoit Morel, GeneRax
Thank you for the quick response.
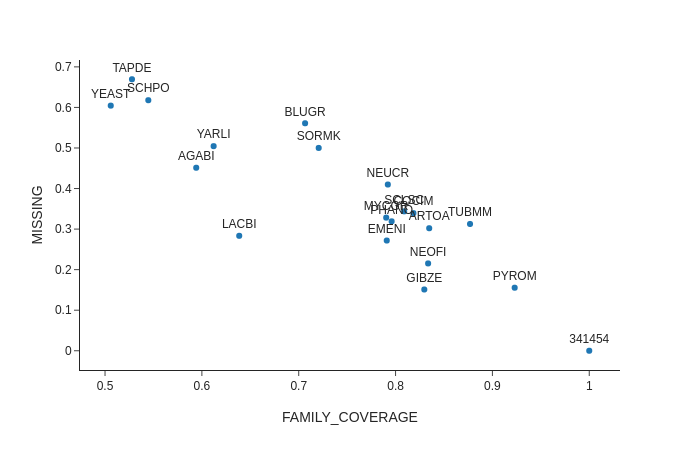
I'm sorry if I wasn't really clear before.
Indeed I did not mean that the values in fractionmissing should sum to 1 but what I meant is that the values in fractionmissing.txt + the values in perSpeciesCoverage.txt (defined in the wiki exactly as you described in the previous mail) for each species should.
I attach a plot so that maybe it is clearer what I mean. On Y there is the value in fractionmissing and on x the value on perSpeciesCoverage. I expected all points to lie in the diagonal 0,1.

(I have generax 2.0.2 compiled from GitHub)
Thanks again!
Giacomo
--
You received this message because you are subscribed to the Google Groups "GeneRax" group.
To unsubscribe from this group and stop receiving emails from it, send an email to generaxusers...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/generaxusers/83c794e2-5961-4540-b536-13e5bceeffcfn%40googlegroups.com.
For more options, visit https://groups.google.com/d/optout.
Benoit Morel
Sep 23, 2021, 4:10:17 AM9/23/21
to GeneRax
All my apologies, my first answer was wrong!
The perSpeciesCoverage.txt is doing what I described.
The fractionMissing.txt is counting the number of gene copies (so after a duplication, a gene will be counted twice) for a given species over all gene families. This number is divided by the the number of gene copies for the species with the highest number of gene copies. And it's then outputting one minus this ratio. So in the end, the species with the highest number of gene copies will have the value 0.
I don't remember why I output this file, I can't think about a good usecase for this weird quantity...
Does it make more sense with your data?
Best,
Benoit
Giacomo Mutti
Sep 23, 2021, 5:42:27 AM9/23/21
to Benoit Morel, GeneRax
Dear Benoit,
Thank you very much for your answer. This makes sense to me!
Thanks again and have a nice day!
Giacomo
To view this discussion on the web visit https://groups.google.com/d/msgid/generaxusers/019b4bbf-d602-4f09-8913-bd2f29e9175fn%40googlegroups.com.
Reply all
Reply to author
Forward
0 new messages
