ssGSEA error
100 views
Skip to first unread message
Yuqin Feng
Jul 3, 2022, 3:02:14 AM7/3/22
to GenePattern Help Forum
Hello everyone,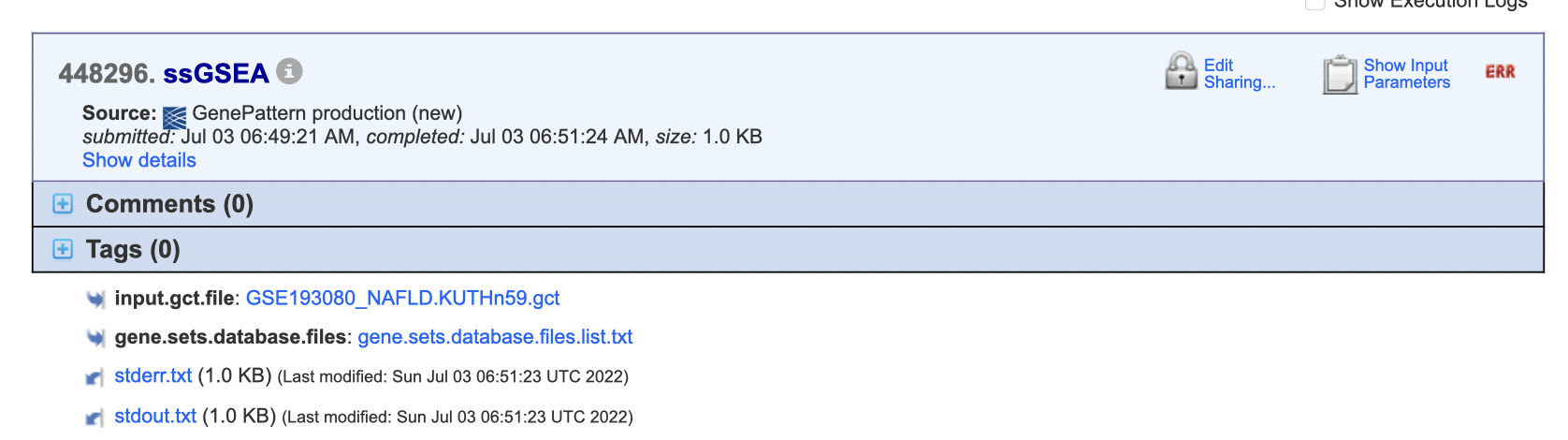
I just tried "ssGSEA" with uploaded gene list GMX file. Job Number:448296. The files were formatted as guidance.

The error was like:
stdout.txt:
WARNING: ignoring environment value of R_HOME
stderr.txt:
Error in matrix("null", nrow = max.N, ncol = max.G) :
invalid 'ncol' value (too large or NA)
Calls: ssGSEA.cmdline
Execution halted
If someone knows what the problem was, thanks a lot in advance!
Anthony Castanza
Jul 4, 2022, 2:49:15 PM7/4/22
to genepatt...@googlegroups.com
Hello,
In the GMX file you provided all of the data is surrounded by quotation marks, The gmx file specification does not support quoted data and the parser used by ssGSEA does not support automatically stripping these characters. Please produce a version of the gmx file without quotes and try it again.
Let me know if you have any questions
-Anthony
Anthony S. Castanza, PhD
Curator, Molecular Signatures Database
Mesirov Lab, Department of Medicine
University of California, San Diego
--
You received this message because you are subscribed to the Google Groups "GenePattern Help Forum" group.
To unsubscribe from this group and stop receiving emails from it, send an email to genepattern-he...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/genepattern-help/c244d36a-222d-4ced-8de7-b545b963d649n%40googlegroups.com.
Yuqin Feng
Jul 11, 2022, 5:02:53 AM7/11/22
to GenePattern Help Forum
Thank you for quick reply!
I tried another version of same file without the quotation marks in Job Number: 449565.But same errors still exist.
Maybe is there other problem?
Anthony Castanza
Jul 11, 2022, 7:48:17 PM7/11/22
to genepatt...@googlegroups.com
Hmm, the only thing I can see is that you have gene_name gene_id for the headers for your first two columns. They should be NAME and Description per the gct spec: https://software.broadinstitute.org/cancer/software/gsea/wiki/index.php/Data_formats#GCT:_Gene_Cluster_Text_file_format_.28.2A.gct.29
I thought the parser in the ssGSEA module was a little more permissive than this, but for the sake of debugging please try changing the header so it's in strict compliance with the specification and give it another try.
-Anthony
Anthony S. Castanza, PhD
Curator, Molecular Signatures Database
Mesirov Lab, Department of Medicine
University of California, San Diego
To view this discussion on the web visit https://groups.google.com/d/msgid/genepattern-help/320f1bf0-5988-4352-b650-34d30e6ee89dn%40googlegroups.com.
Anthony Castanza
Jul 12, 2022, 3:10:52 PM7/12/22
to GenePattern Help Forum
Hello again,
One of my colleagues pointed out that there are still potentially issues with the gmx file. There appears to be a small formatting issue where the new version has an extra header "V1" and "V2" this row should be removed.
Additionally, the second gene set ends with a series of NA genes to fill the gaps between the gene lengths.
I'm not 100% sure that either this or the previous suggestion will correct the error you're observing, but would recommend correcting these two issues in addition to the header formatting of the gct file I mentioned previously and giving it another go.
Please let me know if you're still experiencing issues with this module after these adjustments
-Anthony
Anthony S. Castanza, PhD
Curator, Molecular Signatures Database
Mesirov Lab, Department of Medicine
University of California, San Diego
Reply all
Reply to author
Forward
0 new messages
