test significance and stability of MFA estimates/ statistics based on bootstrap
Drummer
I have a dataset of a cross design experiment with (subjects * stimuli = all subjects go through all stimuli) and I measured 2 groups of variables (3 biological response measures and 2 ratings.
My goal is to fit a MFA that will show how these groups of variables organise/separate dimensionally.
First I fitted this with the MFA functions in FactorMiner
second: I got the eigenvalues for the extracted dimensions, as well as some loadings and metrics that describe the quality of variable and dimension relationships.
I would like to assess the relative stability and significance of these variables and dimensions. So I decided to use bootstrapping to resample with replacement from the original data to create new datasets, and for each iteration (total of 1500) I refitted an MFA/PCA and stored the metrics that I am interested in (eigenvalues for the first dimensions, loadings, etc).
Here is where I get a bit lost. Right now what I have done was to compute 95%CIs in the resampled data (for example eigenvalues for the first component in Figure 1). But not sure this is enough nor how to interpret. For example, the observed eigenvalue was 1.14, I see that the bootstrapped 95%CIs distribution ranges from 1.07 to 1.21, since this is above 1 I would intuitively think this component is reliable.
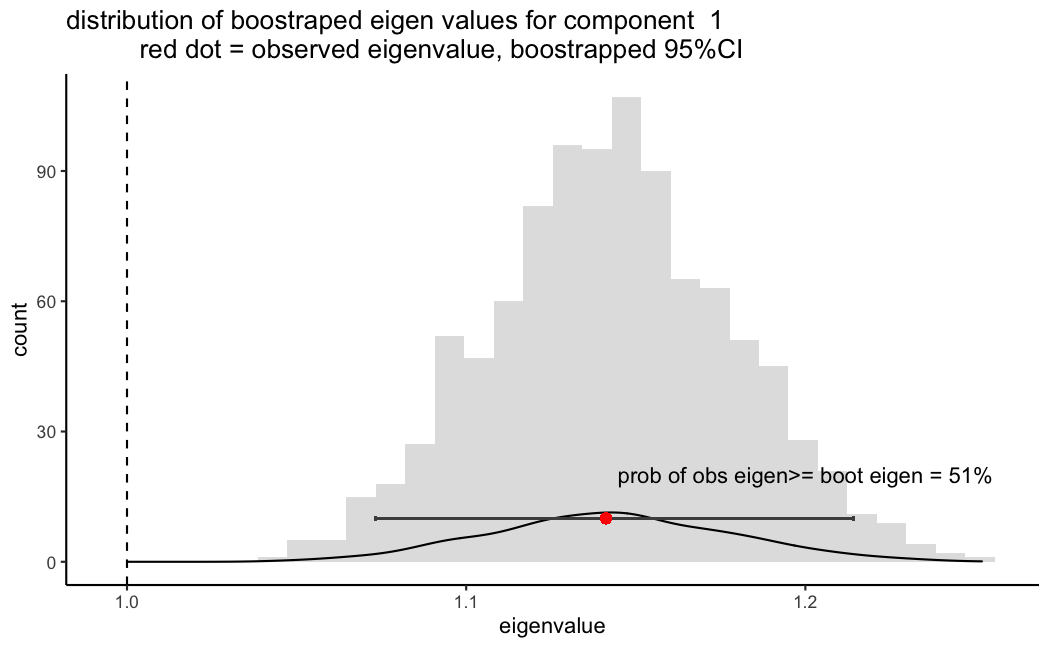
However, some suggested I should compute the probability of obtaining a more extreme eigenvalue than my observed one. Based on the counts though (or just looking at the above figure) this probability (number of eigenvalues equal to or more extreme than the observed/total number of bootstrapped eigenvalues) is about .5 (I am assuming is equivalent to a p.value, thus it would not be significant, or in other words the probability of getting such value by chance is high)
The other thing I read in a few papers was that I should actually compute a difference between my observed eigenvalue - and all individual bootstrap eigenvalues to create a distribution and then derive the 95CIs and test significance of a null of zero (not sure I understand the later) but I did this and you can see and you can see in the figure 2 however, this now includes zero.
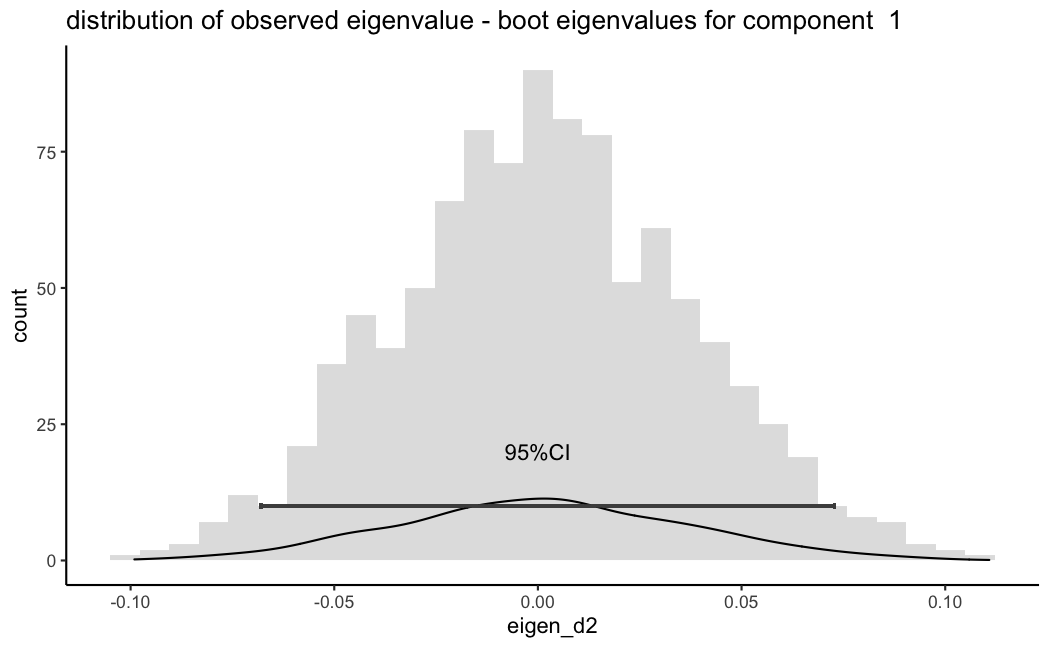
can anyone comment on the appropriateness and correctness of any of these options including whether I misinterpreted this approach or are there any simple alternatives that will allow me to give some confidence around estimates of my MFA/PCA
Thanks for your help
