extractptclfromseg.py command not found
28 views
Skip to first unread message
Himanshu Sharma
Sep 6, 2021, 9:43:11 AM9/6/21
to EMAN2
Hi Folks,
We just updated our Eman2 version to 2.91
"EMAN 2.91 final ( GITHUB: 2021-03-08 11:36 - commit: 81caed2 )
Your Python version is: 3.7.9"
While attempting to extract particles from tomograms, I got an error where the extractptclfromseg.py command was "not found".(command and output mentioned below)
"
$ extractptclfromseg.py Position_15_stack_full_rotx_bin5_preproc__tube_final.hdf Position_15_stack_full_rotx_bin5_preproc.hdf --random 100 --edge 32 --thresh 1.3
$ extractptclfromseg.py: command not found.
"
The functionalities before this step seemed to work well. Not sure why this error shows up? Do I need to install anything additional to run the extraction functionalities?
Best
-Himanshu
Muyuan Chen
Sep 6, 2021, 11:32:27 PM9/6/21
to em...@googlegroups.com
It has been renamed to e2spt_extractfromseg.py. You can also find it through the e2projectmanager GUI.
--
--
----------------------------------------------------------------------------------------------
You received this message because you are subscribed to the Google
Groups "EMAN2" group.
To post to this group, send email to em...@googlegroups.com
To unsubscribe from this group, send email to eman2+un...@googlegroups.com
For more options, visit this group at
http://groups.google.com/group/eman2
---
You received this message because you are subscribed to the Google Groups "EMAN2" group.
To unsubscribe from this group and stop receiving emails from it, send an email to eman2+un...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/eman2/d4610b6b-f23d-42be-ba99-857536b8d30bn%40googlegroups.com.
Himanshu Sharma
Sep 7, 2021, 6:18:17 AM9/7/21
to em...@googlegroups.com
Thanks, Muyuan, this works well now. But, I have another question at this stage.
Using the above command, I am trying to extract particles from a tomogram generated using IMOD. It seems that some of the functionalities as not as straightforward as for tomograms generated in eman2.
Is there a tutorial or source where I can get a workflow for processing these tomograms imported from IMOD??
Best
-h
You received this message because you are subscribed to a topic in the Google Groups "EMAN2" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/eman2/HlNRpi87Ssg/unsubscribe.
To unsubscribe from this group and all its topics, send an email to eman2+un...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/eman2/CAO_0xqGisfOduQOT3PkSTPNGS7aXF_7iPCEEzAS7AVrYom1j0w%40mail.gmail.com.
Himanshu
Research Scholar
Department of Bio-sciences and Bio-engineering
Indian Institute of Technology Guwahati
Assam-781039, India
Research Scholar
Department of Bio-sciences and Bio-engineering
Indian Institute of Technology Guwahati
Assam-781039, India
Alt email : s.him...@iitg.ernet.in
Ludtke, Steven J.
Sep 7, 2021, 8:40:17 AM9/7/21
to em...@googlegroups.com
Hi Himanshu,
see my earlier reply on 4/13/21. Using tomograms created by IMOD means you cannot do per-particle tilt series extraction, which means most of the remaining EMAN2 pipeline cannot be run, so doing this isn't a focus of any of our tutorials. Don't
get me wrong, there is nothing wrong with IMOD tomograms. We used IMOD ourselves for many years. The issue is that we don't have the necessary information to map precisely from the IMOD tomograms back to the tilt series. The particle extraction process normally
extracts BOTH the 3-D particles and the 2-D per particle tilt series simultaneously.
Having said that, it should still be quite possible to do what you are trying to do, but "It seems that some of the functionalities as not as straightforward as for tomograms generated in eman2". Isn't really enough for us to give you assistance.
When you ask for help, including the complete command-line you tried to run, along with either errors you got or an expectation of what you expected vs what you got is the minimum we'd need. It's like calling your mechanic and saying "my car is making a clunking
sound. What's wrong with it?" without letting him see the car or hear the sound.
--------------------------------------------------------------------------------------
Steven Ludtke, Ph.D. <slu...@bcm.edu> Baylor College of Medicine
Steven Ludtke, Ph.D. <slu...@bcm.edu> Baylor College of Medicine
Charles C. Bell Jr., Professor of Structural Biology
Dept. of Biochemistry and Molecular Biology (www.bcm.edu/biochem)
Dept. of Biochemistry and Molecular Biology (www.bcm.edu/biochem)
To view this discussion on the web visit https://groups.google.com/d/msgid/eman2/CAHWT1o4yOwwTxHjx6Ae57HRXUiCzzncgtQmMEbCd89WqhE_xKg%40mail.gmail.com.
MuyuanChen
Sep 7, 2021, 10:25:40 AM9/7/21
to em...@googlegroups.com
My general recommendation is to import IMOD aligned tilt series (and tlt file) instead of tomograms, then run e2tomogram with —niter=0 and —noali. So EMAN will handle the reconstruction and every program in the pipeline shall work. This is not an ideal solution since more interpolation steps happen in the process which may limit the resolution. Simply doing re-run tilt series alignment in EMAN avoid this issue, and should not be much more painful since it is fully automatic.
Muyuan
On Sep 7, 2021, at 5:17 AM, Himanshu Sharma <ace.him...@gmail.com> wrote:
To view this discussion on the web visit https://groups.google.com/d/msgid/eman2/CAHWT1o4yOwwTxHjx6Ae57HRXUiCzzncgtQmMEbCd89WqhE_xKg%40mail.gmail.com.
Himanshu Sharma
Sep 10, 2021, 12:10:44 PM9/10/21
to EMAN2
Thanks, Steve and Muyuan, and I appreciate the suggestions.
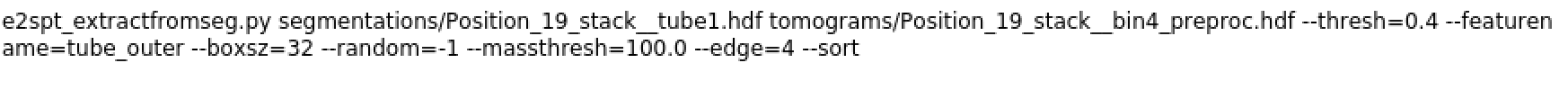
@Steve: Yes I remember your older suggestion as I had raised a similar issue before. Given the same reason, I did not want to throw in the error messages without knowing if a protocol for working on imported tomograms already existed. Anyway. I have been working with my tomograms between IMOD and Eman2. For some reason (could be a problem with my data), the reconstruction does not seem to be equally good in EMAN2, hence I had to resort to importing the tomograms.
Having said this, I again tried to follow the full pipeline in Eman2 and faced the following errors while trying to find particles from segmentation:
command: (I tried varying the box size and massthread but the problem persists)
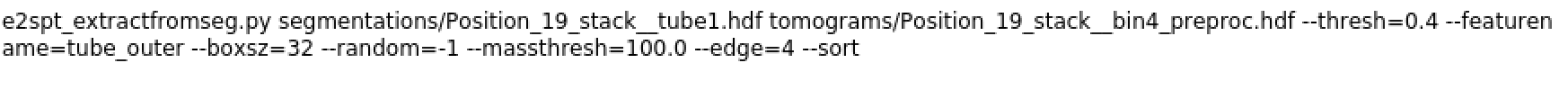
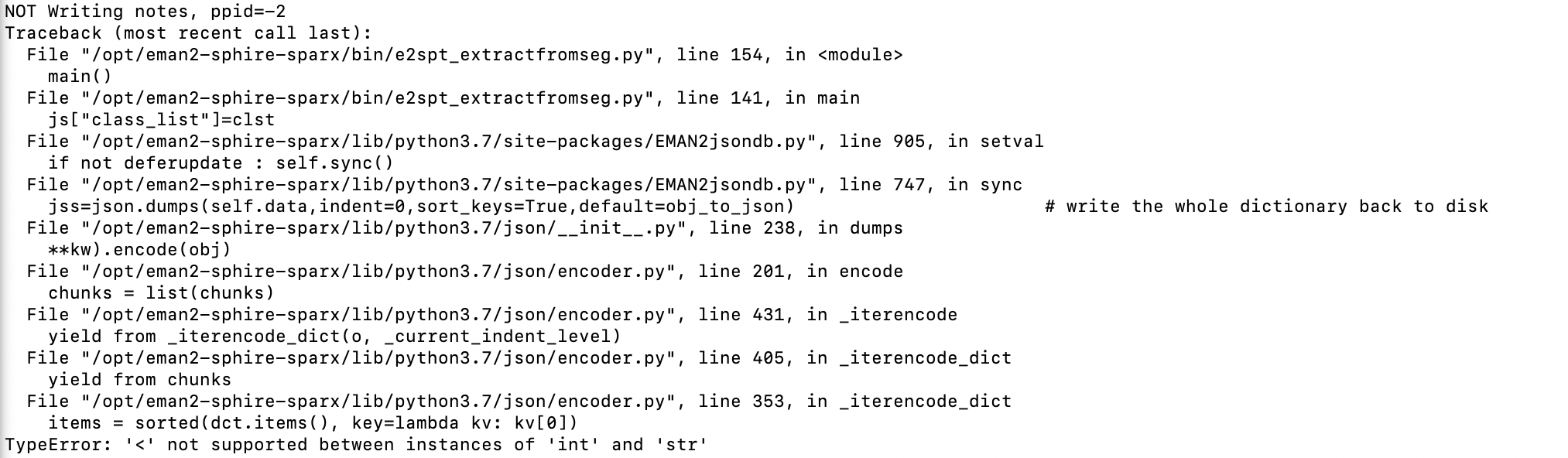
error:

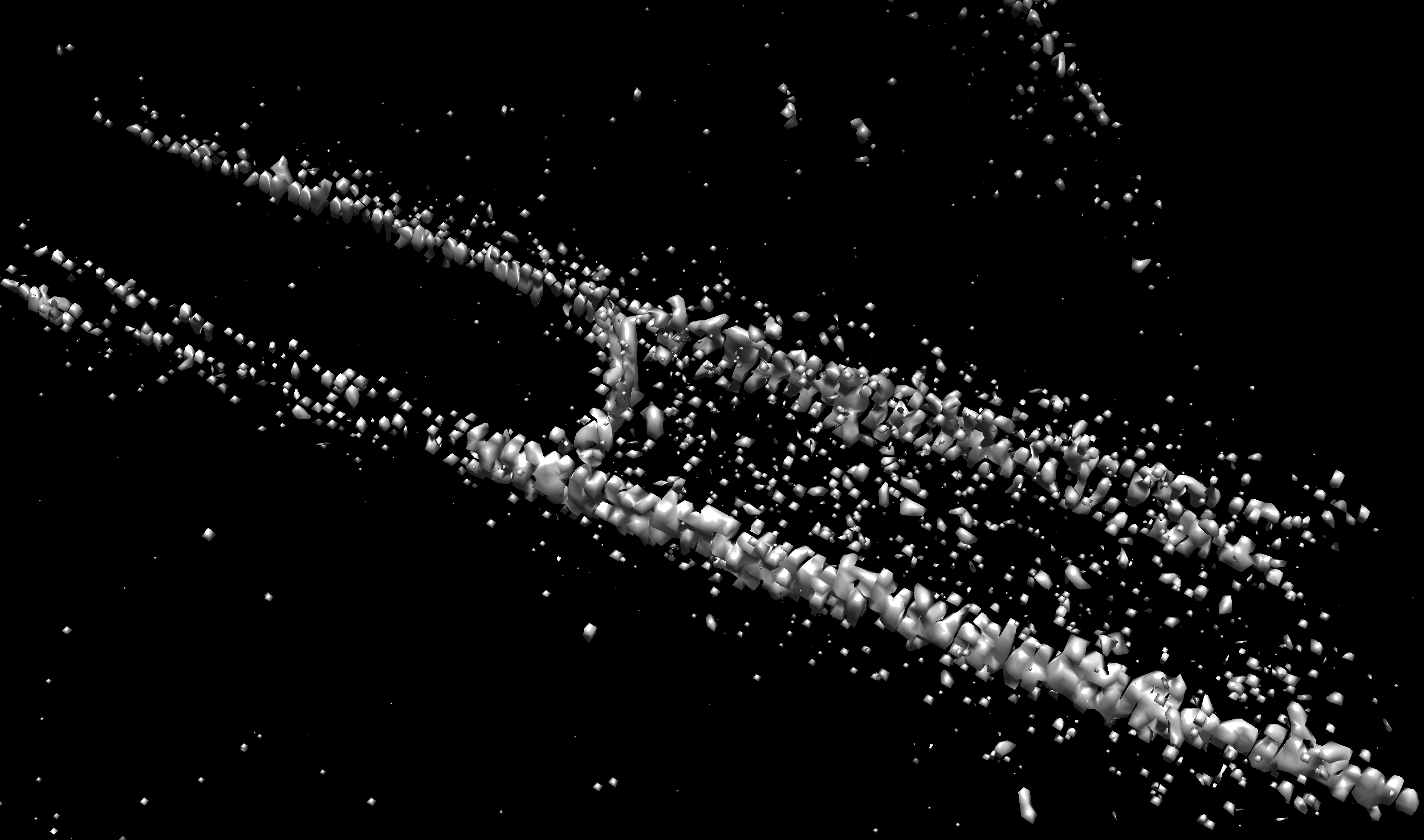
the segmentation does not look great at this stage, but as I am just trying to get an initial test model, I did not bother improving this further. I intend to extract parts of the outer membrane as seen here.

@Muyuan: Importing the aligned tilt series does seem a very interesting suggestion. Do you refer to the *.ali files (and the tlt, is this the rawtilt file?) ?? And if yes, could CTF corrected and dose fractioned files work as well?
Thanks
-Himanshu
MuyuanChen
Sep 10, 2021, 12:46:45 PM9/10/21
to em...@googlegroups.com
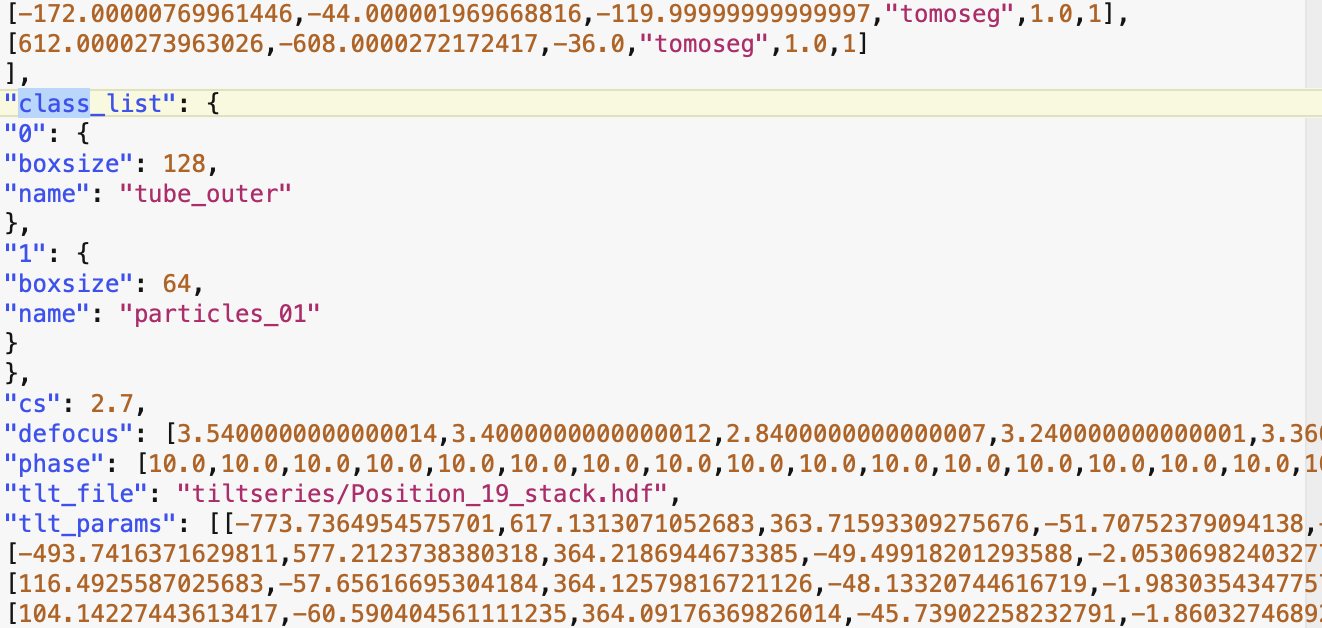
For the e2spt_extractfromseg error, do you trigger this on any tomogram, or just one of them? If you clear all particles selected from a tomogram, then run the program again, do you trigger the same error? My guess is there are some particles labeled with integers instead of strings for some reason. You can also just open the corresponding json file in the info directory and check the “class_list” key.
It would be good to know what exactly went wrong in the EMAN reconstruction so we may be able to improve it. Yes you can use the .ali file and the .tlt from IMOD alignment. Ideally not the .rawtlt since that would be the angle from the microscope before alignment. Then you would at least know whether the difference come from alignment or reconstruction. If you have done ctf correction in IMOD, then just skip the step in EMAN. Otherwise, using the ctf from EMAN is fine too. Not sure how your dose fraction is done, but I don’t think it makes much a difference in general...
On Sep 10, 2021, at 11:10 AM, Himanshu Sharma <ace.him...@gmail.com> wrote:
Thanks, Steve and Muyuan, and I appreciate the suggestions.@Steve: Yes I remember your older suggestion as I had raised a similar issue before. Given the same reason, I did not want to throw in the error messages without knowing if a protocol for working on imported tomograms already existed. Anyway. I have been working with my tomograms between IMOD and Eman2. For some reason (could be a problem with my data), the reconstruction does not seem to be equally good in EMAN2, hence I had to resort to importing the tomograms.Having said this, I again tried to follow the full pipeline in Eman2 and faced the following errors while trying to find particles from segmentation:command: (I tried varying the box size and massthread but the problem persists)
<Screenshot 2021-09-10 at 17.58.57.png>error:
<Screenshot 2021-09-10 at 17.59.24.png>
the segmentation does not look great at this stage, but as I am just trying to get an initial test model, I did not bother improving this further. I intend to extract parts of the outer membrane as seen here.
<Screenshot 2021-09-10 at 18.02.40.png>
To view this discussion on the web visit https://groups.google.com/d/msgid/eman2/379b9d37-484f-4cb9-8059-35f80006c56cn%40googlegroups.com.
<Screenshot 2021-09-10 at 18.02.40.png><Screenshot 2021-09-10 at 17.59.24.png><Screenshot 2021-09-10 at 17.58.57.png>
Himanshu Sharma
Sep 13, 2021, 10:30:10 AM9/13/21
to EMAN2
Hi,
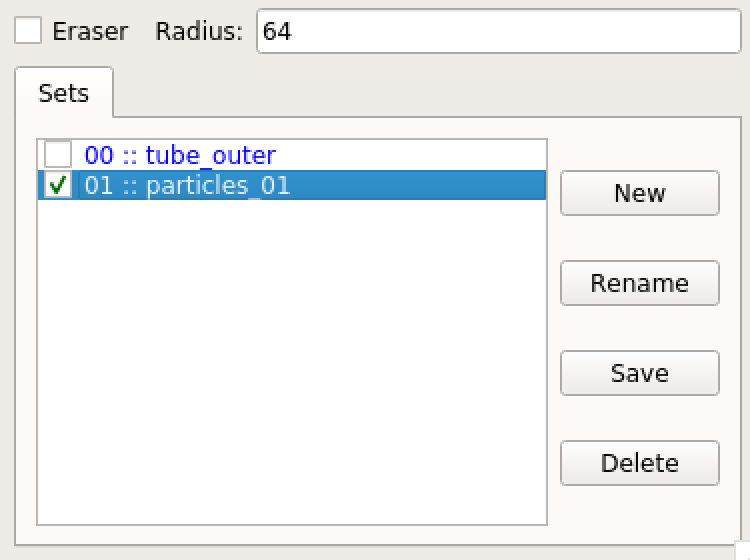
The e2spt_extractfromseg error was seen one more than one tomogram. I have tried this with three tomograms and seem to get this error. I tried to clear all the particles and the error persists. However, there is something I observed after running the command. In spite of the error, the program seems to find some particles (particles_01 in the image) and this (particle identification) does not appear in the program log. Additionally, the identified particles do not seem to be good representatives from the trained model (seem to be random picks at best in high contrast features of the tomogram).

this is a screenshot from the tomgram.json file:

Thanks
MuyuanChen
Sep 13, 2021, 10:58:42 AM9/13/21
to em...@googlegroups.com
I see. This bug was fixed on April 15th, 2021, so it probably still exist in 2.91. That’s probably why I was not able to reproduce it… I took a look at the older version, and you are right that it should not affect the particle selection, only the label of the particles will be set to default.
This program mostly just select high value voxels from the provided segmentation file as particles in the original tomogram. So if the segmentation is not ideal, the particle positions won’t be either. There are many reasons that can cause a non-ideal segmentation, but it is hard to guess from the information here. If you are just selecting particles from tomograms, I am now recommending this https://blake.bcm.edu/emanwiki/EMAN2/e2tomo_more#Automated_particle_selection
The design is similar to the segmentation model, but with an interactive GUI it is easier to figure out what went wrong and adjust the training set to get better results.
Muyuan
On Sep 13, 2021, at 9:30 AM, Himanshu Sharma <ace.him...@gmail.com> wrote:
Hi,The e2spt_extractfromseg error was seen one more than one tomogram. I have tried this with three tomograms and seem to get this error. I tried to clear all the particles and the error persists. However, there is something I observed after running the command. In spite of the error, the program seems to find some particles (particles_01 in the image) and this (particle identification) does not appear in the program log. Additionally, the identified particles do not seem to be good representatives from the trained model (seem to be random picks at best in high contrast features of the tomogram).
<Screenshot 2021-09-13 at 16.18.18.png>this is a screenshot from the tomgram.json file:
To view this discussion on the web visit https://groups.google.com/d/msgid/eman2/82a0eaef-7b91-410b-afef-465eda31a06cn%40googlegroups.com.
<Screenshot 2021-09-13 at 16.25.38.png><Screenshot 2021-09-13 at 16.18.18.png>
Reply all
Reply to author
Forward
0 new messages
