A question about nsym
62 views
Skip to first unread message
Liang Chen
Jul 17, 2022, 11:17:00 PM7/17/22
to EMAN2
Dear EMAN2 Community,
I have been struggling with EMAN2's helical symmetry for a long time. Really need your help.
For example, I am trying to reconstruct a microtubule. The repeating unit rotates 24° and rises 10.67A.
The "tz" will be 10.67A / 9.06 apix = 1.12 pix?
If box size is 72pix, the "nsym" should be 72/1.12 = 64, so take half as 32?
Should the whole symmetry be H32:1:24:1.12?
I tried this number. EMAN2 gives me a really strange shape.
Best Regards,
Liang
Ludtke, Steven J.
Jul 17, 2022, 11:28:00 PM7/17/22
to em...@googlegroups.com
Is that the A/pix value for the downsampled 1k map or the full resolution map? When you do e2spt_refine, you are working with the fully sampled data, not the downsampled data.
You realize microtubules aren't true helices, and have a seam, right?
--------------------------------------------------------------------------------------
Steven Ludtke, Ph.D. <slu...@bcm.edu> Baylor
College of Medicine
Charles C. Bell Jr., Professor of Structural Biology Dept. of Biochemistry
Deputy Director, Advanced Technical Cores and Molecular Biology
Academic Director, CryoEM Core
Co-Director CIBR Center
--
--
----------------------------------------------------------------------------------------------
You received this message because you are subscribed to the Google
Groups "EMAN2" group.
To post to this group, send email to em...@googlegroups.com
To unsubscribe from this group, send email to eman2+un...@googlegroups.com
For more options, visit this group at
http://groups.google.com/group/eman2
---
You received this message because you are subscribed to the Google Groups "EMAN2" group.
To unsubscribe from this group and stop receiving emails from it, send an email to eman2+un...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/eman2/d7c8e84a-9603-4285-a498-2563469def7fn%40googlegroups.com.
Liang Chen
Jul 17, 2022, 11:38:26 PM7/17/22
to EMAN2
I think it is a bin2x map with dimensions "1344, 2046, 101" downloaded as a tutorial sample from IMOD website. I encountered trouble with my own data, so I think I should try other tomograms to make sure the symmetry could work.
Even if microtubules are not true helices, I should get some roughly similar shape, right? When I use "c1", EMAN2 gives me the correct shape with missing wedge. But when I applied "H32:1:24:1.12", the shape totally messed up. Let's assume microtubules possess ideal helical symmetry. Is my "nsym" correct?
Ludtke, Steven J.
Jul 18, 2022, 7:20:53 AM7/18/22
to em...@googlegroups.com
It would be very useful if you could post a screenshot of what you're seeing. "Messed up" could mean anything.
Do you have the helix aligned in the correct orientation? Helical symmetry in EMAN2 is applied along the Z axis. If your object isn't aligned on the Z axis, obviously you will not get what you're hoping for when you symmetrize.
--------------------------------------------------------------------------------------
Steven Ludtke, Ph.D. <slu...@bcm.edu> Baylor
College of Medicine
Charles C. Bell Jr., Professor of Structural Biology Dept. of Biochemistry
Deputy Director, Advanced Technical Cores and Molecular Biology
Academic Director, CryoEM Core
Co-Director CIBR Center
To view this discussion on the web visit https://groups.google.com/d/msgid/eman2/9e1ea784-0e56-4779-87e0-46baa1790f92n%40googlegroups.com.
Liang Chen
Jul 18, 2022, 7:39:52 AM7/18/22
to EMAN2
I think they are aligned along z axis, as I gave EMAN2 a reference cylinder along z axis. Is there such command I could use?
I tried restrict the tilt angle to 10 but the latest version failed with errors, and the old version gave me something weird. Please see the images below. The subtomograms' orientations should be roughly the same.
The image below uses symmetry H32:1:24:1.12, no tilt angle restriciton.

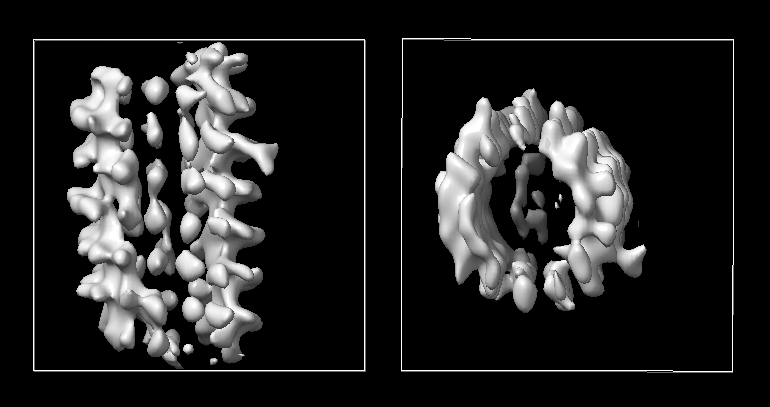
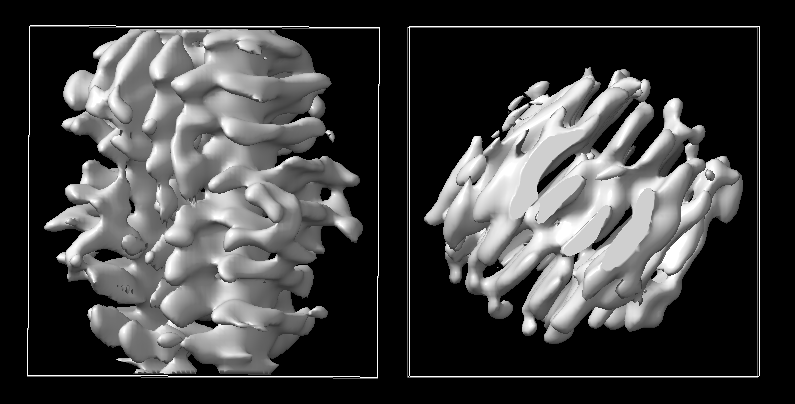

The image below uses c1, no tilt angle restriciton.

The image below uses c1 with tilt angle 10.

Ludtke, Steven J.
Jul 18, 2022, 8:42:49 AM7/18/22
to em...@googlegroups.com
The reconstruction without restrictions looks visually ok, but it's impossible to tell what orientation you are showing us, and you'll note that looking along the helical axis (roughly) in the center right image, that the helix is clearly not centered on the
Z axis. When you impose symmetry, it is absolutely necessary for the object to be in the correct orientation AND centered on the origin. Using a cylindrical reference for the refinement doesn't force the result to be properly centered without any imposed symmetry.
It may tend to align things along the correct axis, but even that isn't guaranteed as it depends on whether the reference has a similar cylindrical radial density profile.
If you impose symmetry during refinement, then the symmetry enforcement enforces the center and orientation, though with a poor reference, it is still possible to wind up with incorrect maps.
Starting with your c1 without tilt restriction map, the first thing to do would be to center the map. You could do this, for example, with e2proc3d.py --process and one of:
xform.center :
xform.centeracf :
xform.centerofmass : int_shift_only(INT) powercenter(INT) threshold(FLOAT)
(from e2help.py processor center)
However, personally, I think I would do this with e2filtertool.py where you can interactively adjust the parameters and see the results:
the xform.applysym processor allows you to impose symmetry as part of the processor chain.
--------------------------------------------------------------------------------------
Steven Ludtke, Ph.D. <slu...@bcm.edu> Baylor
College of Medicine
Charles C. Bell Jr., Professor of Structural Biology Dept. of Biochemistry
Deputy Director, Advanced Technical Cores and Molecular Biology
Academic Director, CryoEM Core
Co-Director CIBR Center
On Jul 18, 2022, at 6:39 AM, Liang Chen <liangche...@gmail.com> wrote:
I think they are aligned along z axis, as I gave EMAN2 a reference cylinder along z axis. Is there such command I could use?
I tried restrict the tilt angle to 10 but the latest version failed with errors, and the old version gave me something weird. Please see the images below. The subtomograms' orientations should be roughly the same.
The image below uses symmetry H32:1:24:1.12, no tilt angle restriciton.
<H32.1.24.1.12.png>The image below uses c1, no tilt angle restriciton.
<c1.png>The image below uses c1 with tilt angle 10.
<tilt10.png>
To view this discussion on the web visit https://groups.google.com/d/msgid/eman2/8e2a62ff-9741-48e4-91fa-18ddf1003e58n%40googlegroups.com.
<tilt10.png><c1.png><H32.1.24.1.12.png>
Liang Chen
Jul 18, 2022, 9:13:40 AM7/18/22
to EMAN2
Thank you for the instructions! The z axis is pointing out of screen in all the right panels above.
Is the symmetry and center option supposed to be imposed after alignment?
The following command doesn't work:
e2proc3d.py spt_00/threed_05.hdf spt_00/threed_05_center_applysym.hdf --process xform.center --process xform.applysym H32:1:24:1.12
How do I let symmetry be imposed while EMAN2 is doing averaging? How to force the filament in the central place while averaging?
Previously, c4 symmetry seems automatically impose central position and applies symmetry for each iteration, but H symmetry never worked for me.
Ludtke, Steven J.
Jul 18, 2022, 10:57:03 AM7/18/22
to em...@googlegroups.com
Again, "doesn't work" isn't generally very useful without the message. However, in this case, the problem is obvious, so I can reply. Try this:
the xform.applysym processor was for use with e2filtertool.py.
e2proc3d.py spt_00/threed_05.hdf spt_00/threed_05_center_applysym.hdf --process xform.center --sym H32:1:24:1.12
While generally anything you can do in filtertool you can also do with e2proc2d or e2proc3d, there is a bug related to helical symmetry, so you have to do it the above way. (explanation: Since the syntax for specifying parameters at the command
line is --process processorname:key=value:key=value the ":" in helical symmetry don't work right in this case. ie - you should have said --process xform.applysym:sym=H32:1:24:1.12 but that wouldn't have worked either due to the ":" required for helical symmetry.)
--------------------------------------------------------------------------------------
Steven Ludtke, Ph.D. <slu...@bcm.edu> Baylor
College of Medicine
Charles C. Bell Jr., Professor of Structural Biology Dept. of Biochemistry
Deputy Director, Advanced Technical Cores and Molecular Biology
Academic Director, CryoEM Core
Co-Director CIBR Center
To view this discussion on the web visit https://groups.google.com/d/msgid/eman2/be352250-adfd-493a-9230-adf07e096acfn%40googlegroups.com.
Reply all
Reply to author
Forward
0 new messages
