ShortBred AMR data
Josie Blair
Lo, Chien-Chi
Hi,
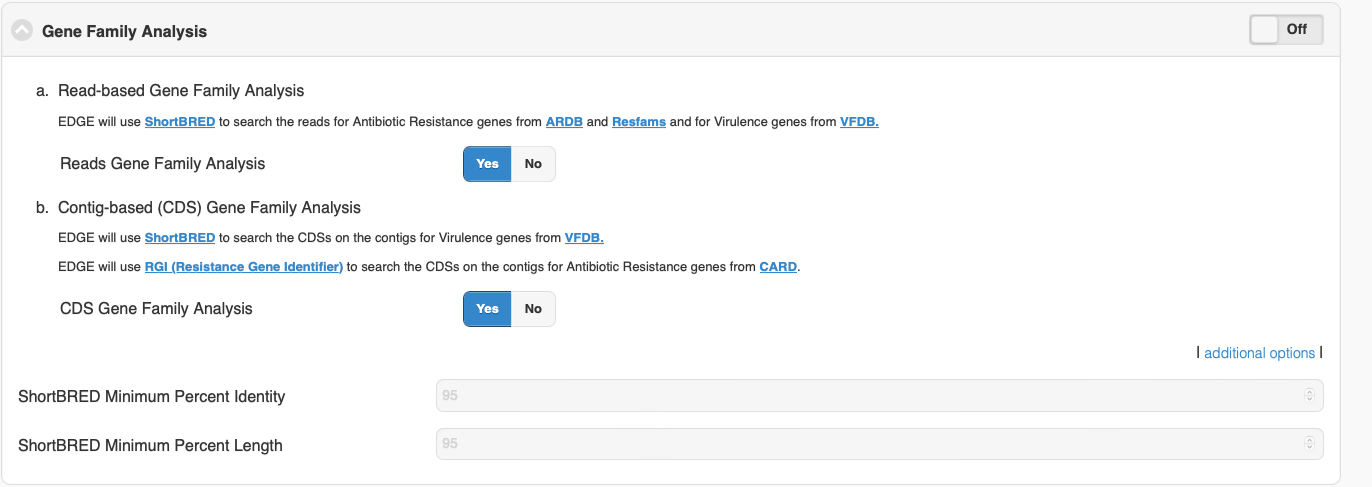
According the my understanding, the ShortBRED is a tool to screen a metagenome or metatranscriptome against a given AMR marker set to profile the presence/absence and relative abundance of the AMR proteins. It based on RAPsearch2 software for fast protein similarity search from short reads. EDGE allows user to adjust default identity and length % threshold (see attached screenshot when user click on addition options). I don’t think ShortBRED will identify mutations in these genes.
Thanks,
Chienchi

--
You received this message because you are subscribed to the Google Groups "edge-users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to
edge-users+...@googlegroups.com.
To view this discussion on the web visit
https://groups.google.com/d/msgid/edge-users/787730e9-d917-4805-a860-b61b6cf3ec26n%40googlegroups.com.
Lo, Chien-Chi
Hi,
Just another thoughts on your question. Since your data is from Nanopore sequencing. The quality of the raw reads may not good enough for SNP/mutations detection and it may have many false variants call. EDGE can assemble nanopore reads and use contig-based gene family analysis which uses RGI to search the AMR genes from CARD. The result of RGI does report SNPs.
Thanks,
Chienchi
To view this discussion on the web visit
https://groups.google.com/d/msgid/edge-users/CF188EF2-74C9-45BE-B71D-EE957C7BC5D0%40lanl.gov.
Josie Blair
On Jan 18, 2023, at 3:36 PM, Lo, Chien-Chi <chie...@lanl.gov> wrote:
Hi,
Just another thoughts on your question. Since your data is from Nanopore sequencing. The quality of the raw reads may not good enough for SNP/mutations detection and it may have many false variants call. EDGE can assemble nanopore reads and use contig-based gene family analysis which uses RGI to search the AMR genes from CARD. The result of RGI does report SNPs.
Thanks,
Chienchi
From: "'Lo, Chien-Chi' via edge-users" <edge-...@googlegroups.com>
Reply-To: Chien-Chi Lo <chie...@lanl.gov>
Date: Tuesday, January 17, 2023 at 2:27 PM
To: Josie Blair <josie...@gmail.com>, edge-users <edge-...@googlegroups.com>
Subject: Re: [EXTERNAL] ShortBred AMR data
Hi,
According the my understanding, the ShortBRED is a tool to screen a metagenome or metatranscriptome against a given AMR marker set to profile the presence/absence and relative abundance of the AMR proteins. It based on RAPsearch2 software for fast protein similarity search from short reads. EDGE allows user to adjust default identity and length % threshold (see attached screenshot when user click on addition options). I don’t think ShortBRED will identify mutations in these genes.
Thanks,
Chienchi