Visualize the fiber tracts generated from MRtrix
434 views
Skip to first unread message
Z S
Sep 17, 2022, 2:17:36 PM9/17/22
to DSI Studio
Hi Frank,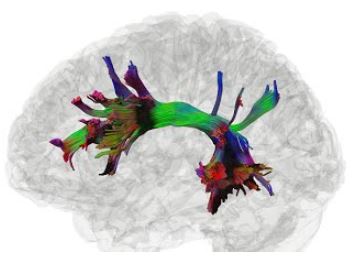

Recently, I generated fiber tracts from MRtrix and I wanted to show them in DSI Studio (like this).
I tried to learn this visualization from your video. https://www.youtube.com/watch?v=PdjfjRW7bLw
Step1, I converted the tck file to trk file.
Step2, I loaded the FA map and T1W map into DSI Studio, ran registration.
Step3, I loaded the trk map into DSI Studio. But, I got the following map. So, we can see the brain and fiber tracts are separated.
Do you have any thoughts about this issue?
Thanks,
Zhiqiang
Fang-Cheng Yeh
Sep 17, 2022, 2:56:21 PM9/17/22
to dsi-s...@googlegroups.com
How did you convert from tck to trk?
You may need to shift the coordinates to align the image space.
Frank
--
You received this message because you are subscribed to the Google Groups "DSI Studio" group.
To unsubscribe from this group and stop receiving emails from it, send an email to dsi-studio+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/96602622-b745-4e77-aa84-0fe817418b7bn%40googlegroups.com.
Z S
Sep 18, 2022, 5:45:00 AM9/18/22
to DSI Studio
Hi Frank,
Thanks for your suggestions. Get it. I need to use FA.nii.gz as a reference map, rather than T1.nii.gz. Now all is well.
Best,
Zhiqiang
Katja Heuer
Oct 1, 2023, 5:15:42 AM10/1/23
to DSI Studio
Hi Frank and Zhiqiang,
I have a similar issue (conversion failed, and loading the tck directly shows them in a different space). Please, could you let me know how you converted from tck to trk? Would you be happy to share the code / command line with me?
Thank you so much in advance.
Have a lovely Sunday!
Best,
Katja
Frank Yeh
Oct 3, 2023, 4:31:16 PM10/3/23
to katja...@gmail.com, DSI Studio
Hi Katja,
The solution depends on the space the tck was created in.
Is it possible to upload a sample data (tck and fib.gz) so that I can look into the issue?
Best,
Frank
On Sun, Oct 1, 2023 at 5:15 AM Katja Heuer <katja...@gmail.com> wrote:
Hi Frank and Zhiqiang,I have a similar issue (conversion failed, and loading the tck directly shows them in a different space). Please, could you let me know how you converted from tck to trk? Would you be happy to share the code / command line with me?Thank you so much in advance.Have a lovely Sunday!Best,Katja
On Sunday, 18 September 2022 at 11:45:00 UTC+2 Z S wrote:
Hi Frank,Thanks for your suggestions. Get it. I need to use FA.nii.gz as a reference map, rather than T1.nii.gz. Now all is well.Best,Zhiqiang
在2022年9月17日星期六 UTC+2 20:56:21<Frank Yeh> 写道:
How did you convert from tck to trk?You may need to shift the coordinates to align the image space.FrankOn Sat, Sep 17, 2022 at 2:17 PM Z S <zhiq...@gmail.com> wrote:
Hi Frank,Recently, I generated fiber tracts from MRtrix and I wanted to show them in DSI Studio (like this).
I tried to learn this visualization from your video. https://www.youtube.com/watch?v=PdjfjRW7bLwStep1, I converted the tck file to trk file.Step2, I loaded the FA map and T1W map into DSI Studio, ran registration.Step3, I loaded the trk map into DSI Studio. But, I got the following map. So, we can see the brain and fiber tracts are separated.
Do you have any thoughts about this issue?Thanks,Zhiqiang
--
You received this message because you are subscribed to the Google Groups "DSI Studio" group.
To unsubscribe from this group and stop receiving emails from it, send an email to dsi-studio+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/96602622-b745-4e77-aa84-0fe817418b7bn%40googlegroups.com.
--
You received this message because you are subscribed to the Google Groups "DSI Studio" group.
To unsubscribe from this group and stop receiving emails from it, send an email to dsi-studio+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/ba49c061-02ab-4d98-93d1-19d96baae9a5n%40googlegroups.com.
Reply all
Reply to author
Forward
0 new messages
