Iteratively applying multiple ROIs for subject specific network analysis from ommand-line
72 views
Skip to first unread message
William McCuddy
Jun 3, 2021, 10:54:32 PM6/3/21
to DSI Studio
Hello,
Does anyone have an example command for iteratively applying pairs of ROIs from a list of 17 for a total 136 ROI-ROI contingencies. The 17 ROIs have been selected from the HCP-MP Atlas based on their overlap with the default mode network. The goal is to automatically generate tracts comprising the DMN. I might also need to add in some ROAs to keep every tidy, but I can probably figure that out later.
These are the specific ROIs
1 L_10r
2 L_a24
3 L_p32
4 L_s32
5 L_31a
6 L_31pd
7 L_31pv
8 L_d23ab
9 L_POS1
10 L_POS2
11 L_RSC
12 L_v23ab
13 L_IP1
14 L_PFm
15 L_PGi
16 L_PGs
17 L_TPOJ3
Thanks!
Travis
Frank Yeh
Jun 3, 2021, 11:19:12 PM6/3/21
to dsi-s...@googlegroups.com
First make sure you have 17 regions saved as one nii.gz file (DSI
Studio should also have its region labels as nii.gz.txt)
Then the following command should work out for you:
dsi_studio --action=trk --source=subject.fib.gz --fiber_count=1000000
--output=no_file --connectivity=my_HCP_MMP.nii.gz
--connectivity_value=trk --connectivity_type=pass
Best regards,
Frank
> --
> You received this message because you are subscribed to the Google Groups "DSI Studio" group.
> To unsubscribe from this group and stop receiving emails from it, send an email to dsi-studio+...@googlegroups.com.
> To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/57fea5bb-01d9-420f-8fb4-77286bfbae9cn%40googlegroups.com.
Studio should also have its region labels as nii.gz.txt)
Then the following command should work out for you:
dsi_studio --action=trk --source=subject.fib.gz --fiber_count=1000000
--output=no_file --connectivity=my_HCP_MMP.nii.gz
--connectivity_value=trk --connectivity_type=pass
Best regards,
Frank
> You received this message because you are subscribed to the Google Groups "DSI Studio" group.
> To unsubscribe from this group and stop receiving emails from it, send an email to dsi-studio+...@googlegroups.com.
> To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/57fea5bb-01d9-420f-8fb4-77286bfbae9cn%40googlegroups.com.
William McCuddy
Jun 4, 2021, 12:24:05 AM6/4/21
to DSI Studio
Thanks for quick response! I tried running the cmd in a bash script (see below) and replaced [mysubject] and [my_HCP_MMP] with my specific file names. The script runs, but i get all 1000000 streamlines when i open the resulting .trk file in the gui. I made sure to have all 17 regions saved as one nii.gz file (which i did via Regions-->Save all regions as... I'm able to open this file in the gui and see all 17 ROIs displayed). The nii.gz.txt appears to be fine as well. I wasn't sure if these files needed to be save in the dsi-studio folder or the subject folder (where the .fib file is saved). I tried saving copies in both places and got the same result. I also tried to make another nii.gz file with just two regions and I got the same result (.trk file showing all 100000 streamlines).
This is what the simple script looks like:
path=D:\Imaging dsi_studio64
dsi_studio --action=trk --source=[mysubject].fib.gz --fiber_count=1000000
--output=no_file --connectivity=[my_HCP_MMP].nii.gz
--connectivity_value=trk --connectivity_type=pass
--connectivity_value=trk --connectivity_type=pass
Any idea what I'm doing wrong?
Frank Yeh
Jun 4, 2021, 12:37:34 AM6/4/21
to dsi-s...@googlegroups.com
It must be a problem in DSI Studio.
Could you add > log.txt at the end of your command and post the
content of log.txt output?
I will fix it as soon as possible.
Frank
> To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/bd99f795-9f78-41ed-9d93-8e686283ea7dn%40googlegroups.com.
Could you add > log.txt at the end of your command and post the
content of log.txt output?
I will fix it as soon as possible.
Frank
William McCuddy
Jun 4, 2021, 6:56:31 AM6/4/21
to DSI Studio
Here is output of cmd line (changed SubjectFib to deidentify):
D:\DICOMOBJ\[SubjectFib]>--output=no_file --connectivity=L_DMN.nii.gz
'--output' is not recognized as an internal or external command,
operable program or batch file.
D:\DICOMOBJ\
[SubjectFib] >--connectivity_value=trk --connectivity_type=pass
'--connectivity_value' is not recognized as an internal or external command,
operable program or batch file.
D:\[SubjectFib]>log.txt
Here is output of text file:
DSI Studio Jul 1 2020, Fang-Cheng Yeh
source=sample.fib.gz
action=trk
loading sample.fib.gz...
sample.fib.gz does not exist. terminating...
It feels like I'm doing something wrong (likely), but DSI knows SubjectFib.gz exists because i get a SubjectFib.gz.trk.gz output.
Thanks for your help!
Frank Yeh
Jun 4, 2021, 8:22:40 AM6/4/21
to dsi-s...@googlegroups.com
Could you update DSI studio and try again?
Frank
To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/5c9ab805-487e-4b2b-b309-64d8797eb8e5n%40googlegroups.com.
William McCuddy
Jun 4, 2021, 8:44:51 AM6/4/21
to DSI Studio
Stupid question. To update, do i need to download latest version (and if so, is this the appropriate link for download:http://dsi-studio.labsolver.org/dsi-studio-download) or is there a way to update from the cmd line?
Thanks
Frank Yeh
Jun 4, 2021, 8:52:06 AM6/4/21
to dsi-s...@googlegroups.com
Sorry it cannot be updated from the command line. You would need to
get it from the download link you mentioned.
Frank
> To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/ceaa02f1-5a3a-4b43-a40e-fbd850e4f323n%40googlegroups.com.
get it from the download link you mentioned.
Frank
William McCuddy
Jun 4, 2021, 9:40:57 AM6/4/21
to DSI Studio
Updated (GUI says "June 2 2021 build")
I generated new src file and fib file, re-ran command, and got same result except now I get .tt.gz file
Output of log.txt:
D:\[path to subject folder containing new .fib] >path=D:\Imaging\dsi_studio_64
D:\[path to subject folder containing new .fib] >dsi_studio --action=trk --source=D:\
[path to subject folder containing new .fib]\my subject.fib.gz --fiber_count=1000000
D:\[path to subject folder containing new .fib] >--output=no_file --connectivity=L_DMN.nii.gz
'--output' is not recognized as an internal or external command,
operable program or batch file.
D:\
[path to subject folder containing new .fib] >--connectivity_value=trk --connectivity_type=pass
'--connectivity_value' is not recognized as an internal or external command,
operable program or batch file.
D:\
[path to subject folder containing new .fib] >log.txt
----------
both L_DMN.nii.gz and L_DMN.nii.gz.txt are located in the main DSI studio folder and the subject folder containing fib.gz and .bat file.
Frank Yeh
Jun 4, 2021, 10:08:36 AM6/4/21
to dsi-s...@googlegroups.com
You have to put all commands in the same line...(except for the first
path= xxxx)
Frank
> To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/8771ab8f-2274-4ffc-9033-8213a91f3bden%40googlegroups.com.
path= xxxx)
Frank
William McCuddy
Jun 4, 2021, 10:29:11 AM6/4/21
to DSI Studio
oh, duh! That worked. Thank you helping me figure this out. It is now creating a .tt.gz file for each region ROI. Will it automatically generate a merged file? if not could i add another cmd line to the .bat file?
The closest thing i can find in online documents is:
cluster: run track clustering after fiber tracking. 4 parameters separated by comma are needed: --cluster=METHOD_ID,CLUSTER_COUNT,RESOLUTION,OUTPUT FILENAME
METHOD_ID: 0=single-linkage clustering 1=k means 2=EM
CLUSTER_COUNT: The total number of clusters. In k-means or EM, this is the total number of clusters assigned. In single-linkage, it is the maximum number of clusters allowed to avoid over-segmentation (the remaining small clusters will be group in the last clusters).
RESOLUTION: only used in single-linkage clustering. The value is the mini meter resolution for merging the clusters.
Frank Yeh
Jun 4, 2021, 10:41:33 AM6/4/21
to dsi-s...@googlegroups.com
No, it doesn't. You can merge them using GUI.
I am sorry that the command line does not have a track merging function.
Frank
> To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/5388fea3-cd8e-4d25-8cdc-5dc57381db58n%40googlegroups.com.
I am sorry that the command line does not have a track merging function.
Frank
William McCuddy
Jun 4, 2021, 10:43:38 AM6/4/21
to DSI Studio
Looks like I spoke too soon. While it did create individual tract files for each ROI pair, when opening each tract file there was still over 100000 streamlines for every single pair (instead of showing only those tracts that pass through the two ROIs for each pair). Any idea why?
William McCuddy
Jun 4, 2021, 10:47:44 AM6/4/21
to DSI Studio
this is the command, all in single line (except for the first line with Path to dsi studio, which is not shown)
dsi_studio --action=trk --source=D:\DICOMOBJ\Sub001\401_DTI_32dir_3iso\Sub001\Sub001.src.gz.odf.gqi.1.25.fib.gz --fiber_count=1000000 --output=no_file --connectivity=L_DMN.nii.gz --connectivity_value=trk --connectivity_type=pass > log.txt
Frank Yeh
Jun 4, 2021, 10:53:15 AM6/4/21
to dsi-s...@googlegroups.com
The track numbers aren't necessarily wrong.
You may open the tt.gz file in Step T3 Fiber tracking to see if the
tract did pass through the designated ROI.
Best regards,
Frank
> To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/7af103ab-35a4-47b9-b39f-6510388c764en%40googlegroups.com.
You may open the tt.gz file in Step T3 Fiber tracking to see if the
tract did pass through the designated ROI.
Best regards,
Frank
William McCuddy
Jun 4, 2021, 11:06:12 AM6/4/21
to DSI Studio
Thanks for help and your patience, still just trying to figure this thing out.
Below is pic of 2 rois with manual tracking (left) and the same 2 rois tracted with cmd line (right). The picture on the right is how all tracts generated from paired ROIs in cmd line look.
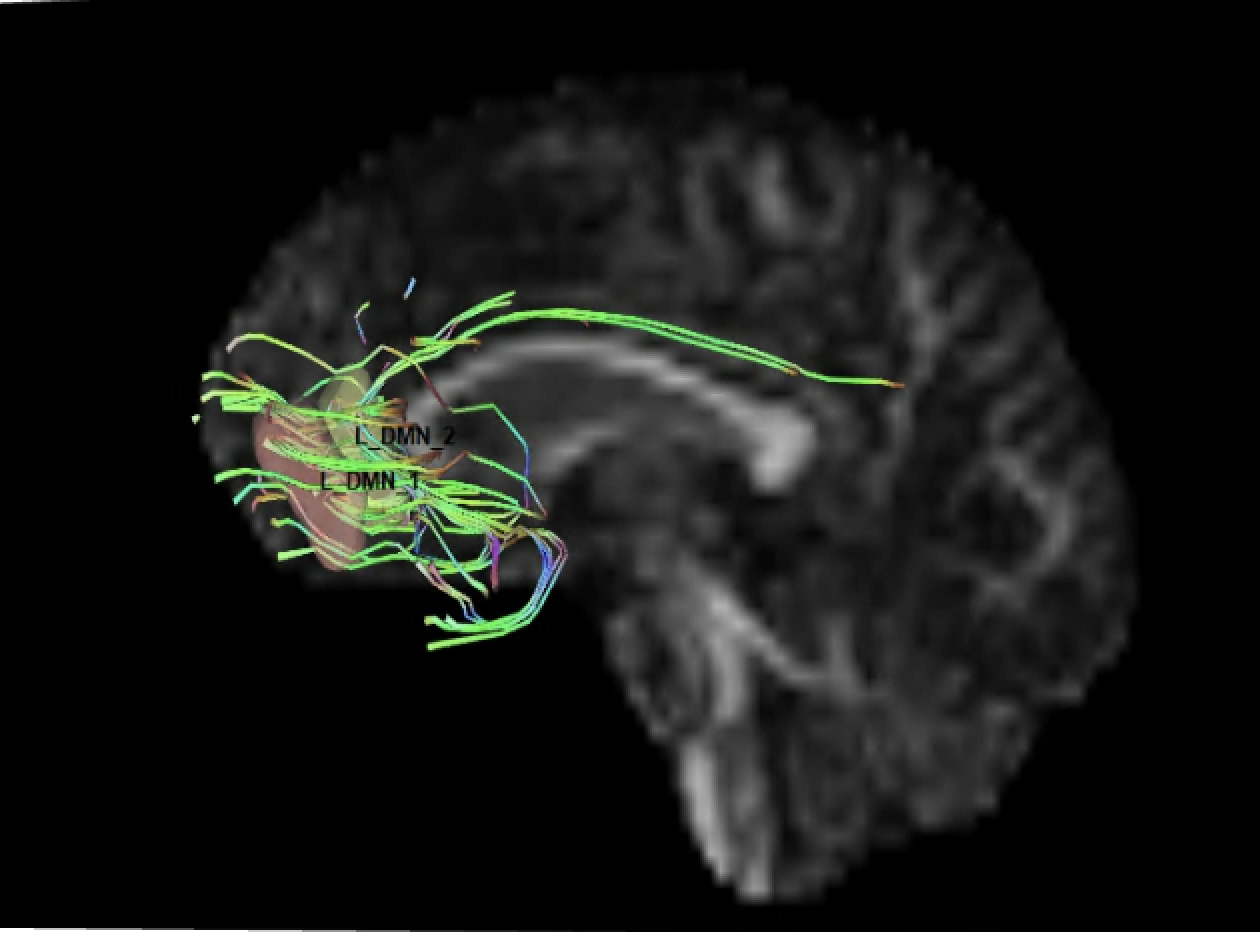
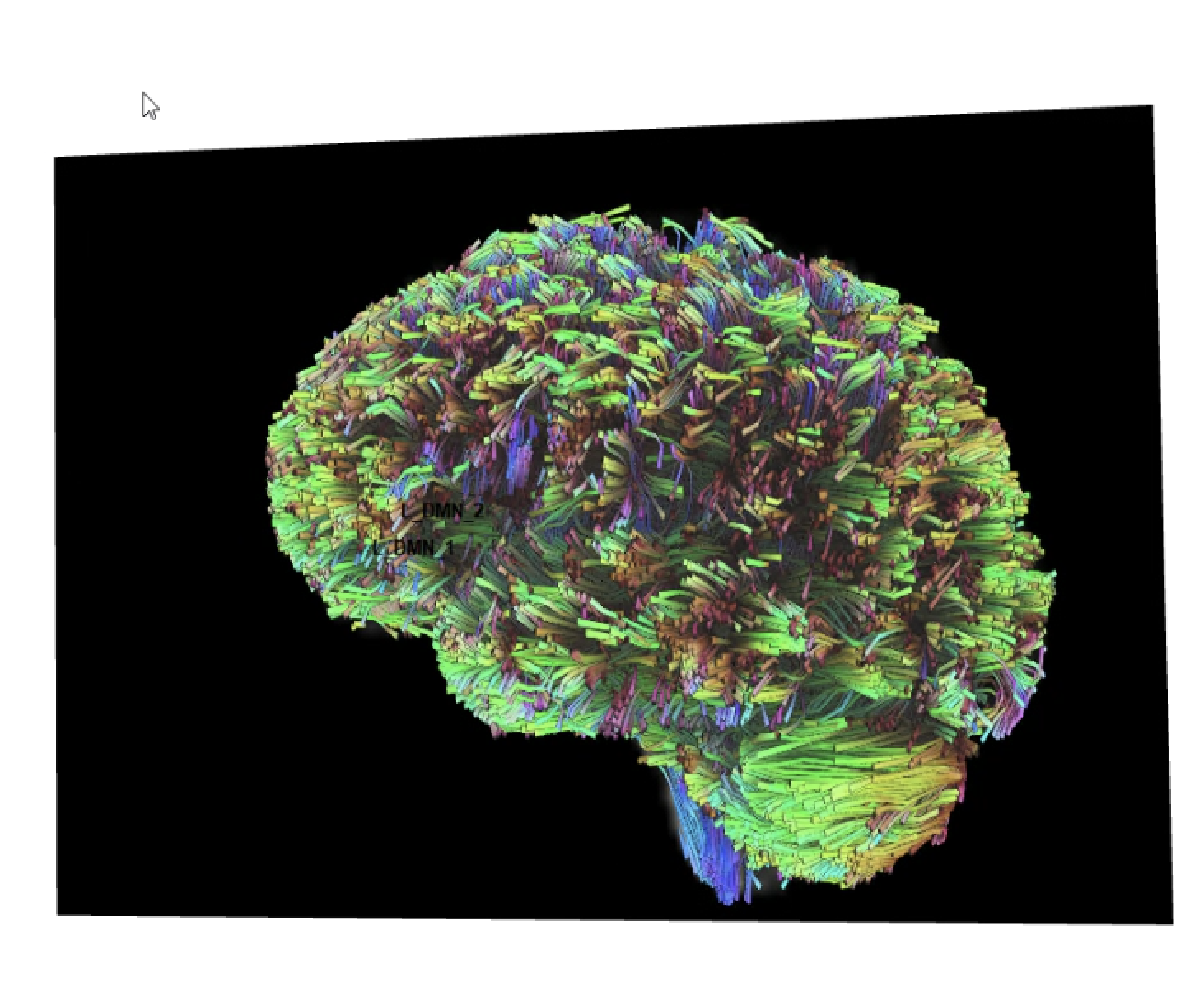
Frank Yeh
Jun 4, 2021, 11:15:24 AM6/4/21
to dsi-s...@googlegroups.com
Could you post the log.txt here?
To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/9de983e6-3459-4fce-aae9-dc087cb1bbbbn%40googlegroups.com.
William McCuddy
Jun 4, 2021, 11:35:49 AM6/4/21
to DSI Studio
It is giving me the same errors as before. I edited the directory names to remove subject/patient names so i could post exact log.txt ([OldDirectoryName]=Sub001). However, even though i also edited the path in .bat file (below), the log.txt file still shows old directory names. I also changed [OldFileName]...fib.gz to Sub001...fib.gz
Cmd line from .bat file (minus first line/path to dsi_studio):
dsi_studio --action=trk --source=D:\DICOMOBJ\Sub001\401_DTI_32dir_3iso\Sub001\Sub001.src.gz.odf.gqi.1.25.fib.gz --fiber_count=1000000 --output=no_file --connectivity=L_DMN.nii.gz --connectivity_value=trk --connectivity_type=pass > log.txt
log.txt output:
D:\DICOMOBJ\[OldDirectoryName]\401_DTI_32dir_3iso\[OldDirectoryName]>path=D:\Imaging\dsi_studio_64
D:\DICOMOBJ\[OldDirectoryName]\401_DTI_32dir_3iso\[OldDirectoryName]>dsi_studio --action=trk --source=D:\DICOMOBJ\[OldDirectoryName]\401_DTI_32dir_3iso\
[OldDirectoryName]\[OldFileName].fib.gz --fiber_count=1000000
D:\DICOMOBJ\[OldDirectoryName]\401_DTI_32dir_3iso\[OldDirectoryName]>--output=no_file --connectivity=L_DMN.nii.gz
'--output' is not recognized as an internal or external command,
operable program or batch file.
D:\DICOMOBJ\[OldDirectoryName]\401_DTI_32dir_3iso\[OldDirectoryName]>--connectivity_value=trk --connectivity_type=pass
'--connectivity_value' is not recognized as an internal or external command,
operable program or batch file.
D:\DICOMOBJ\[OldDirectoryName]\401_DTI_32dir_3iso\[OldDirectoryName]>log.txt
Frank Yeh
Jun 4, 2021, 11:54:05 AM6/4/21
to dsi-s...@googlegroups.com
You will need to make sure that all parameters are in the same line of the command.
There seems to be a line break that cuased the problem.
Frank
To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/083ccab6-e705-4c98-833a-1042cb12ec87n%40googlegroups.com.
Frank Yeh
Jun 4, 2021, 11:56:17 AM6/4/21
to dsi-s...@googlegroups.com
You may need to use a different text editor that surely won't automatically break a line into multiple lines.
Also, check log.txt output.
Otherwise, your command line won't work correctly.
Frank
William McCuddy
Jun 4, 2021, 12:24:40 PM6/4/21
to DSI Studio
confirmed text is all in one line by expanding window (pic below). still same error. I'm going to start from scratch in new directory to see if issue is reproduced.

William McCuddy
Jun 4, 2021, 12:53:52 PM6/4/21
to DSI Studio
OK, starting from scratch at least gave me a log.txt file that makes more sense (below), but same issue as before (one ROI pair tt.gz files contains tracts of whole brain instead of only the tracts that pass through the 2 rois; as seen in pic) Note. I abbreviated the region and label file (L_DMN_test) to contain only 4 regions while testing.
log.txt output:
DSI Studio Jun 2 2021, Fang-Cheng Yeh
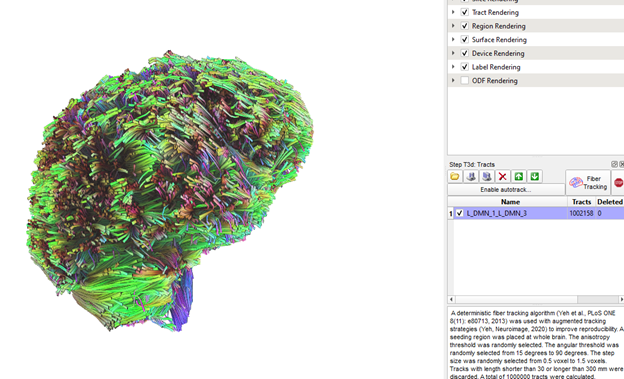
source=D:\DICOMOBJ\Test\Sub001.src.gz.odf.gqi.1.25.fib.gz
action=trk
loading D:\DICOMOBJ\Test\Sub001.src.gz.odf.gqi.1.25.fib.gz...
otsu_threshold=0.6
fa_threshold=0
dt_threshold=0.2
turning_angle=0
step_size=0
smoothing=0
min_length=30
max_length=300
method=0
initial_dir=0
interpolation=0
seed_plan=0
random_seed=0
check_ending=0
tip_iteration=0
fiber_count=1000000
seed_count=0
thread_count=4
start tracking.
thread_count=4
finished tracking.
1000000 tracts are generated using 2109424 seeds.
0 tracts are removed by pruning.
The final analysis results in 1000000 tracts.
output=no_file
connectivity=L_DMN_test.nii.gz
connectivity_type=pass
connectivity_value=trk
loading L_DMN_test.nii.gz
DWI dimension=(128,128,55)
NIFTI dimension=(128,128,55)
nifti loaded as multiple ROI file
looking for region label file
label file loaded:D:/DICOMOBJ/Test/L_DMN_test.txt
a total of 4 regions are loaded.
connectivity_threshold=0.001
count tracks by passing
calculate matrix using trk
saving L_DMN_1_L_DMN_2.tt.gz
saving L_DMN_1_L_DMN_3.tt.gz
saving L_DMN_1_L_DMN_4.tt.gz
saving L_DMN_2_L_DMN_3.tt.gz
saving L_DMN_2_L_DMN_4.tt.gz
saving L_DMN_3_L_DMN_4.tt.gz

William McCuddy
Jun 4, 2021, 1:11:27 PM6/4/21
to DSI Studio
I think I am getting close to figuring this out. The original command you gave me did not have --roi parameter. In the cmd line I added "-roi=L_DMN_test.nii.gz" between --fiber and --output.
Looking at online documentation the following is provided: "(e.g. --roi=nifti.gz:1 only load regions with value=1)" What does the "1 only load regions with value=1" mean?
Frank Yeh
Jun 4, 2021, 1:46:52 PM6/4/21
to dsi-s...@googlegroups.com
Thank you for your help.
I finally found the bug and fixed it.
Please update DSI Studio again (dated today, 1:43 pm) and the updated version should generate a much smaller tt.gz file.
If not, please let me know.
Sorry for the hassle!
Frank
To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/8151e812-cb7b-45bf-86f3-48b3ef750450n%40googlegroups.com.
William McCuddy
Jun 4, 2021, 3:12:51 PM6/4/21
to DSI Studio
Thanks Frank, this is definitely better, but it automatically generated tracts between RO1 and ROI2 still significantly outnumber the manually generated tracts between the same two ROIs (see pics below). Are the standard tracking parameters different when running cmd line vs gui?
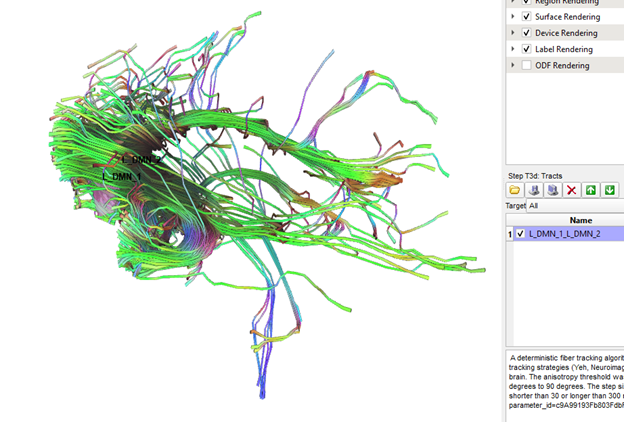
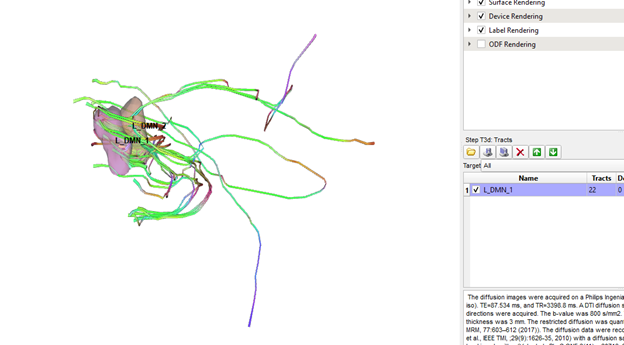
Also, no hassle at all! This is a really cool program and I am very appreciative of all the time and energy you have put into it!


Frank Yeh
Jun 4, 2021, 3:22:26 PM6/4/21
to dsi-s...@googlegroups.com
Yes, there are differences.
The default seed/tract count at [Step T3(c) Options][Tracking Parameters][Terminate If] is only 50000, but the one assigned at --fiber_count is 1000000
To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/a50841e5-17d6-4d30-aad2-887520902bf6n%40googlegroups.com.
Reply all
Reply to author
Forward
0 new messages
