Convert Volume from Dream3D to Neper
170 views
Skip to first unread message
Janzen Choi
Jun 25, 2022, 12:23:40 PM6/25/22
to dream3d-users
Hello,
I want to convert a volume generated in Dream3D into a tessellation in Neper, so that I can produce a tetrahedral mesh of the volume with smooth grain boundaries. I am planning to do so by exporting information from Dream3D, and using it to generate a Neper tessellation file with extension '.tess'.
Could anyone provide some help regarding this?
Thank you very much.
Michael Jackson
Jun 28, 2022, 2:16:34 PM6/28/22
to Janzen Choi, dream3d-users
What input file types does Neper accept then we can try and figure out a way forward.
--
Mike Jackson
--
You received this message because you are subscribed to the Google Groups "dream3d-users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to dream3d-user...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/dream3d-users/25ab6389-b4d9-4865-ba62-16556e9c2e0an%40googlegroups.com.
Joe Wendorf
Jun 28, 2022, 3:38:46 PM6/28/22
to Michael Jackson, Janzen Choi, dream3d-users
Hello Janzen,
I use neper to tessellate and mesh Dream3D datasets. I use a simple MATLAB script to write a .tesr file using h5read() and fprintf(), then I use neper to convert that into a .tess and then .mesh file with the -T and -M modules.
The format for .tesr is located here: https://neper.info/doc/fileformat.html#raster-tessellation-file-tesr
It is a simple plain text format. Everything in [square brackets] is optional. If you leave them blank, neper will calculate it from other parameters or generate random data (ex. random grain orientations).
Regards,
Joe
To view this discussion on the web visit https://groups.google.com/d/msgid/dream3d-users/427D56B5-E7D5-4B36-8D6C-D27A79D912B4%40bluequartz.net.
Janzen Choi
Jun 30, 2022, 3:01:08 AM6/30/22
to dream3d-users
Hello Joe,
Thank you very much for your response. It was extremely helpful.
For reference, I ended up doing this:
1) Add the 'Export INL File' filter to the pipeline to obtain the feature ids corresponding to each voxel.
2) Write a Python script to create a .tesr file with the obtained feature ids.
3) Use Neper to convert the .tesr file into a .tess file, then into a .mesh file.
Regards,
Janzen
Divyajyoti Nayak
Nov 28, 2022, 2:58:45 AM11/28/22
to dream3d-users
Hello Janzen,
Could you share the command you used in neper to convert the .tesr file to a .tess file?
Thanks
Divyajyoti
Janzen Choi
Nov 28, 2022, 8:30:12 PM11/28/22
to dream3d-users
Hello Divyajyoti,
I used the following command.
`neper -T -loadtess <tessellation_file> -format tesr -tesrsize <volume_length> -tesrformat ascii`.
This command loads in a generated tessellation of name, '<tessellation_file>'. It then converts the tessellation into a raster tessellation, with each edge of the volume containing '<volume_length>' voxels. The '-tesrformat ascii' simply stores the IDs of the voxels as readable integers, instead of in binary.
Hope this helps.
Regards,
Janzen
Janzen Choi
Nov 29, 2022, 5:17:50 AM11/29/22
to dream3d-users
Hello Divyajyoti,
I realise that I had read your question wrong.
Yes you can. I used the following command.
'neper -T -n from_morpho -morpho "tesr:file(<raster_tessellation>.tesr)"'
From my experience, the conversion from raster tessellation into a tessellation is a bit finnicky. I have attached an image below of this conversion with a microstructure I did a few months ago.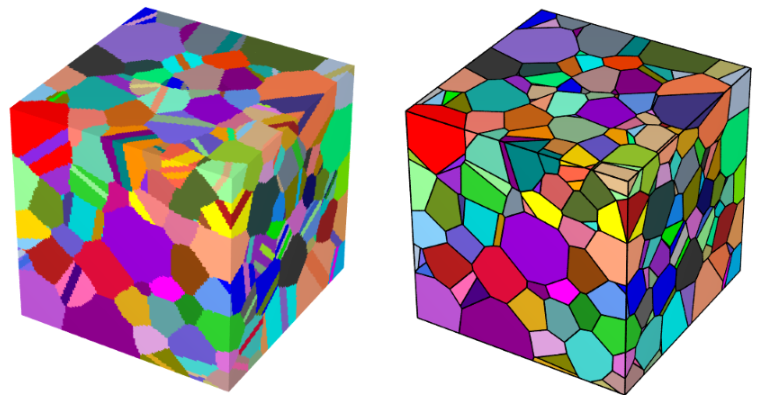 .
.
Specifically, my microstructure had annealling twin structures, which are essentially very thin slices that separated the parent grains. In the conversion, the two sides of the parent grains were basically trying to merge, causing this error.
I also found that you will need a very high resolution to maintain very thin structures. Otherwise, when you convert them into a tessellation, those thin structures will just disappear.
Hope this helps.
Regards,
Janzen
Reply all
Reply to author
Forward
0 new messages
