Post-stratification in the context of a control/treatment experimental design
fernanda...@gmail.com
I'm using DS in a the context of a field experiment that follows a before-after-control-impact statistical design. I want to test whether the deployment of a treatment known to have a positive impact on the breeding output of a bird species could be detected using DS.
I'm just starting to analyse data for the first year of this experiment and I have a question about post-stratification. I'm using the DS package in R, in my data the levels of my Region.Label are "treatmend and control", but I'm looking to get density estimates for survey period: before-control, after-control, before-treatment and after-treatment. So, I post-stratified using the variable survey period as my strata and stratification = "replicate". However, DS undestands my survey effort to be higher than it actually is. I visited all sites 3 times, so survey effort should be 117, but DS is actually considering each level of my survey period and giving the survey effort of 468. Am I missing something here, did I ask for the post stratification using the worng parameters?
Any help would be much appreciated.
Thanks,
Fernanda Alves
Eric Rexstad
the Distance package, help us understand the estimates you wish to obtain. Figures are useful in understanding the design of your experiment.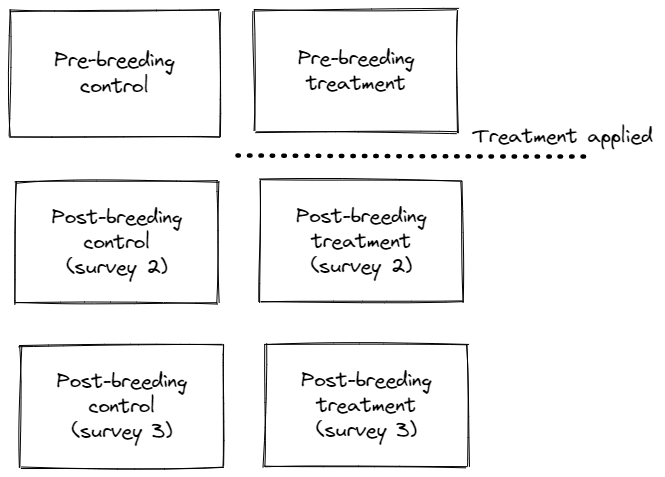
Sent: 15 December 2022 08:42
To: distance-sampling <distance...@googlegroups.com>
Subject: {Suspected Spam} [distance-sampling] Post-stratification in the context of a control/treatment experimental design
You received this message because you are subscribed to the Google Groups "distance-sampling" group.
To unsubscribe from this group and stop receiving emails from it, send an email to distance-sampl...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/distance-sampling/f9436844-9e34-474f-bec3-cf5f5bab2ea1n%40googlegroups.com.
Fernanda Alves
Eric thank you very much for your detailed reply. I really appreciate it.
Yes, the depiction is accurate (thanks for drawing it), and I do have enough detections to fit separate detection functions for each one of the 3 rounds of surveys, which would give me estimates for the 2 levels of my Region.Label (i.e. control/treatment sites). That’s what you mean right? Not 6 detection functions (i.e. one for each treatment type within a round of survey)?
Answering your question about how I expect the treatment to manifest. I expect the effect to be persistent (affecting both 2 and 3). In that case, should I fit the detection function for survey 2 and 3 combined, and do post-stratification? Or would you still simply fit a separate detection function for each round of survey?
PS: Thanks for sending that reference, I’ll look into that approach as well.
Thanks,
Fernanda.
___ ___ ___
{o,o} {-.-} {0,0}
|)__) |)_(| (__(|
-"-"- -"-"- -"-"-
Eric Rexstad
Sent: 16 December 2022 06:53
To: Eric Rexstad <Eric.R...@st-andrews.ac.uk>
Cc: distance-sampling <distance...@googlegroups.com>
Subject: Re: {Suspected Spam} [distance-sampling] Post-stratification in the context of a control/treatment experimental design
Joanne Potts
Eric Rexstad
Sent: 20 March 2023 05:37
To: distance-sampling <distance...@googlegroups.com>
Subject: [distance-sampling] Post-stratification in the context of a control/treatment experimental design
Stephen Buckland
Yes, that is correct. Jo, more details below, which will be more than most will want to know!
You can consider how a count at a given point would be converted to a density estimate. By dividing the point count by the effective area, you get estimated density. Another way to view this is to divide the point count by estimated probability of detection, which gives an estimate of abundance at the circle of radius w about that point, where w is the truncation distance – detected animals beyond w are not included in the count. Then to get from that abundance estimate to a density estimate, you divide by the area of the circle. At least this is the case when the point is surveyed just once. If it is visited say t times, you would divide the count by t, to get the mean count per visit.
The issue now is that we don’t have a good error model for the estimated densities at each point, but we do have suitable models for counts – e.g. Poisson or negative binomial. So instead of taking the estimated density as the response, we take the count, and put the terms to convert it to a density estimate onto the right hand side of the equation model, as a so-called offset. This only works if we us a GLM with a log link function - and the offset is actually the log of the terms taken onto the RHS, and is included in the exponent: E(n)=exp(linear predictor + offset)). In most applications, the offset would be known, but we only have an estimate of it. We handled that by propagating the uncertainty in estimating probability of detection through to the count model, using a bootstrap. Mark Bravington came up with a more sophisticated and less computer-intensive way to achieve the same thing. Or you can be Bayesian, and estimate the offset along with the count model parameters in a single step.
Steve
To view this discussion on the web visit https://groups.google.com/d/msgid/distance-sampling/DBAPR06MB6694677B03AA794562AEE66EEA809%40DBAPR06MB6694.eurprd06.prod.outlook.com.
