Fitting Uniform key without adjustment terms to simulate Strip transectc
Adrian Schiavini
Eric Rexstad
Adrian
Correct that the Distance GUI can be made to 'fit' a flat detection function, whereas Distance in R cannot. I presume you are trying to produce an abundance estimate, and in the situation where detectability is perfect, your abundance estimate is simply number seen scaled up by A/a (ratio of study area size to size of covered region), which you could code yourself.
The mildly tricky part is producing a measure of precision. In
the situation you describe, there would be no uncertainty in the
parameters of the detection function (because there is no
detection function), so you need only the encounter rate
variance. This can be computed using the varn() function inside
the mrds package.
Pretend I have 20 strip transects, all of length 4 units, with detection out to truncation distance 1. Size of covered region is then 160 units. Suppose my study area is of size 1000 units (I cover 16% of the study area with my transects). I fabricate the number of detections per transect as:
nvec=c(0,6,2,4,7,2,1,8,3,13,15,16,10,9,7,3,2,6,3,0)
such that there were 117 detections. My abundance estimate is 117 * (1000/160) = 731.25
varn() can calculate the cv^2 of encounter rate as:
> tmp <- mrds:::varn(lvec=rep(x = 4,times=20), nvec=c(0,6,2,4,7,2,1,8,3,13,15,16,10,9,7,3,2,6,3,0), type="R2") > tmp [1] 0.07180099
making variance of encounter rate
0.392
Variance of my abundance estimate is this encounter rate variance scaled by the A/a multiplier squared (delta method)
> (1000/160)^2 * 0.392
[1] 15.3125
Analytical normal-approximation CI would then be
> sum(nvec)*(1000/160) + c(-1, +1) * 1.96 * sqrt((1000/160)^2 * 15.3125)
[1] 683.3143 779.1857
Somebody on the list will set us right if I've messed up.
--
You received this message because you are subscribed to the Google Groups "distance-sampling" group.
To unsubscribe from this group and stop receiving emails from it, send an email to distance-sampl...@googlegroups.com.
To post to this group, send email to distance...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/distance-sampling/fa2230e5-ec54-4ce1-b776-05e8dfc51902%40googlegroups.com.
For more options, visit https://groups.google.com/d/optout.
-- Eric Rexstad Research Unit for Wildlife Population Assessment Centre for Research into Ecological and Environmental Modelling University of St. Andrews St. Andrews Scotland KY16 9LZ +44 (0)1334 461833 The University of St Andrews is a charity registered in Scotland : No SC013532
Don Carlos
Here is what I did:
nvec=c(0,6,2,4,7,2,1,8,3,13,15,16,10,9,7,3,2,6,3,0)
var.er <- mrds:::varn(lvec=lvec, nvec=nvec, type="R2")
se.er <- sqrt(var.er)
cv.er <- se.er/(sum(nvec)/(sum(lvec)))
A = 1000
a = 160
# abundance estimate
sum(nvec) * (A/a)
# Variance of abundance estimate - delta method
var.a <- (A/a)^2 * cv.er
# Analytical normal-approximation CI
sum(nvec)*(A/a) + c(-1, +1) * 1.96 * sqrt((A/a)^2 * var.a)
Referenced post:
https://groups.google.com/g/distance-sampling/c/QYQ4Oysf9JY/m/UPwlPHYuBwAJ
Eric Rexstad
Don Carlos
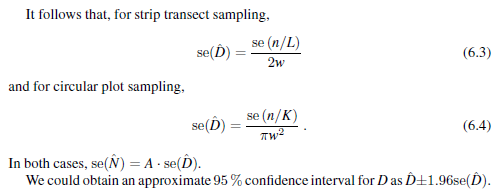
I must apologise. There were errors in my posting from 2017. I've revisited the subject from first principles, based upon the discussion of precision of strip transects found in Buckland et al. (2015):


The only difficult component to calculate is Eqn 6.1. the function `varn` computes the square of this quantity (or variance of encounter rate). Here is the code snippet that performs the calculations:
w <- 1 # strip transect with
half-width=1
lvec <- rep(x = 4,times=20)
nvec <- c(0,6,2,4,7,2,1,8,3,13,15,16,10,9,7,3,2,6,3,0)
var.er <- mrds:::varn(lvec=lvec, nvec=nvec, type="R2")
seD <- sqrt(var.er) / 2*w # se(D-hat) Eqn 6.3
A <- 1000 # study area size
a <- sum(lvec) * 2 * w # covered area
# abundance estimate
abund.est <- sum(nvec) * (A/a)
seN <- seD * A # se(N-hat)
# Analytical normal-approximation CI
normal.ci <- abund.est + c(-1,1)* 1.96 * seN
To view this discussion on the web visit https://groups.google.com/d/msgid/distance-sampling/197091a7-8f25-46a0-9cc3-f9d794367e9bn%40googlegroups.com.
-- Eric Rexstad Centre for Ecological and Environmental Modelling University of St Andrews St Andrews is a charity registered in Scotland SC013532
Don Carlos
mean.s <- mean(df$size, na.rm=T)
# se.s <- sqrt(var(df$size, na.rm=T))
se.s <- sqrt(var(df$size, na.rm=T)/no_of_cluster_detections)
cv.s <- sqrt(var(df$size, na.rm=T)/mean.s)
se.D <- D.hat * sqrt(cv.er^2 + cv.pa^2 + cv.s^2)
# sqrt(D.hat^2*(cv.er^2 + cv.pa ^2 + cv.s^2)) # equivilant, p. 120
cv.D <- se.D / D.hat
Stephen Buckland
For both your se and cv, you seem to have the denominator inside the square root. It should not be.
Steve Buckland
From: distance...@googlegroups.com [mailto:distance...@googlegroups.com]
On Behalf Of Don Carlos
Sent: 30 October 2020 13:32
To: Eric Rexstad <Eric.R...@st-andrews.ac.uk>
Cc: distance-sampling <distance...@googlegroups.com>
Subject: Re: [distance-sampling] Encounter rate variance and normal-approximation CI
Dear Eric,
Thank you very much for the example and scan of the relevant pages - unfortunately, the book is so expensive, so it probably the only one I do not have.
I am actually trying to go through the various calculations of an analysis with a detection function. I am getting most parts of it - based on comparison to the R summary output, but cannot figure out how cluster size (Expected cluster size) and ist se and cv are worked out. Based on the Distance::dht2 function I have done the following:
# calculate varianceand cv for "expected' clusters
mean.s <- mean(df$size, na.rm=T)
# se.s <- sqrt(var(df$size, na.rm=T))
se.s <- sqrt(var(df$size, na.rm=T)/no_of_cluster_detections)
cv.s <- sqrt(var(df$size, na.rm=T)/mean.s)
Although the mean is similar to what I get from the summary outputs in R, the se.s and cv.s differ significantly, although they are extracted from the Distance::dht2 code and seem to follow Buckland et al. 2000, p.120
D.hat <- no_of_cluster_detections *(1/Pa)*mean.s/((2*trunc.dist)*sum(transects$effort))
se.D <- D.hat * sqrt(cv.er^2 +
cv.pa^2 + cv.s^2)
# sqrt(D.hat^2*(cv.er^2 +
cv.pa ^2 + cv.s^2)) # equivilant, p. 120
cv.D <- se.D / D.hat
Thanks so much for all your support on this listserv, much appreciated.
Rgds,
Don
On Fri, 30 Oct 2020 at 14:25, Eric Rexstad <er...@st-andrews.ac.uk> wrote:
Don Carlos
I must apologise. There were errors in my posting from 2017. I've revisited the subject from first principles, based upon the discussion of precision of strip transects found in Buckland et al. (2015):
To view this discussion on the web visit https://groups.google.com/d/msgid/distance-sampling/CAFTQVoCA5G%2B259k4OcdYkcP_fLeUoxw_PRnaBfjiNKgz87mnmg%40mail.gmail.com.