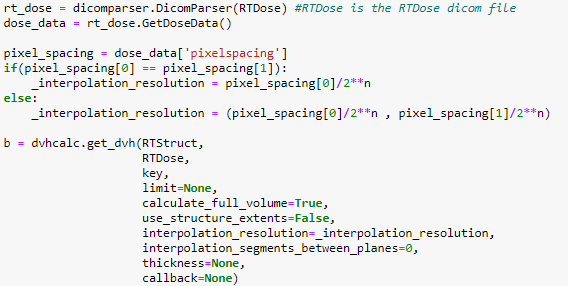
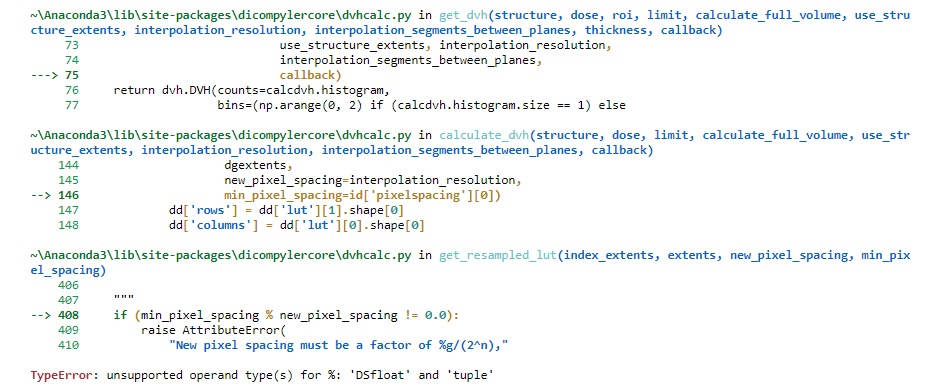
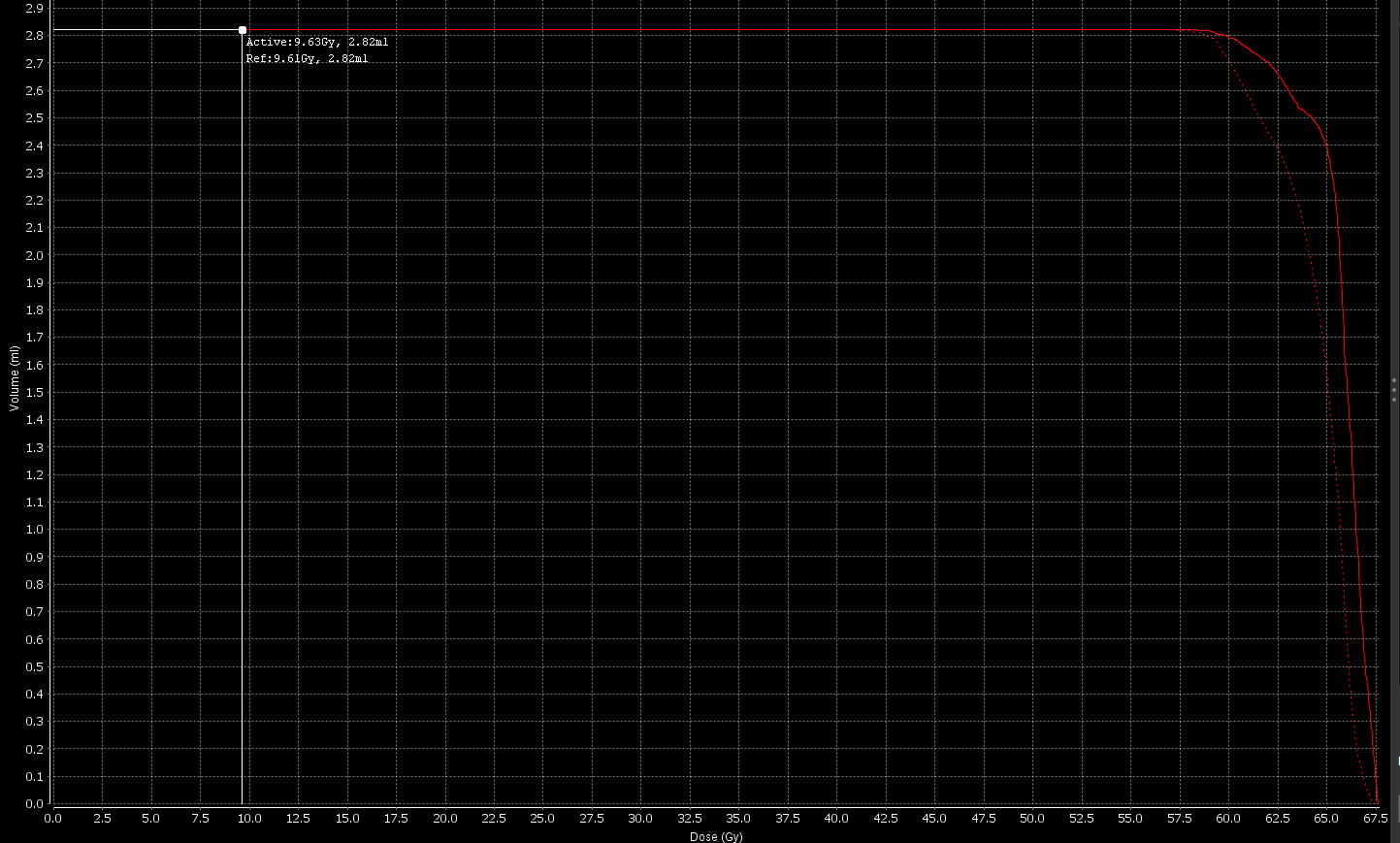
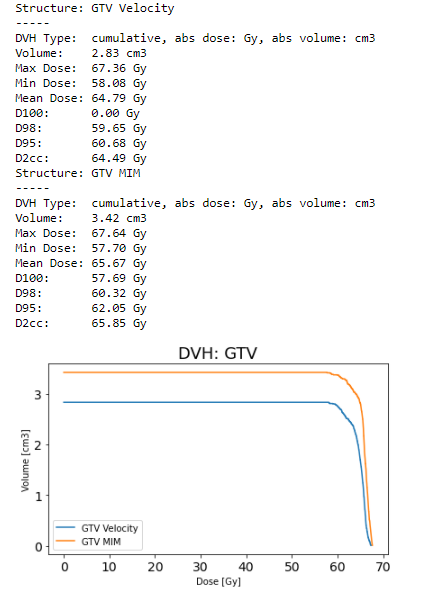
Absolute volume DVHs differ between MIM and dicompyler
Isak Wahlstedt
Dan Cutright
--
-- You received this message because you are subscribed to the Google Groups dicompyler group. To post to this group, send email to dicom...@googlegroups.com. To unsubscribe from this group, send email to dicompyler+...@googlegroups.com. For more options, visit this group at https://groups.google.com/d/forum/dicompyler?hl=en
---
You received this message because you are subscribed to the Google Groups "dicompyler" group.
To unsubscribe from this group and stop receiving emails from it, send an email to dicompyler+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/dicompyler/931eb9b8-76cb-4075-9dca-363f6d1c41f6n%40googlegroups.com.
Aditya Panchal
To view this discussion on the web visit https://groups.google.com/d/msgid/dicompyler/CAMd1neu%3DimG4OqQb075HOdoLf1EgwziyceoPQk3Y95CSmdnQnQ%40mail.gmail.com.
Isak Wahlstedt
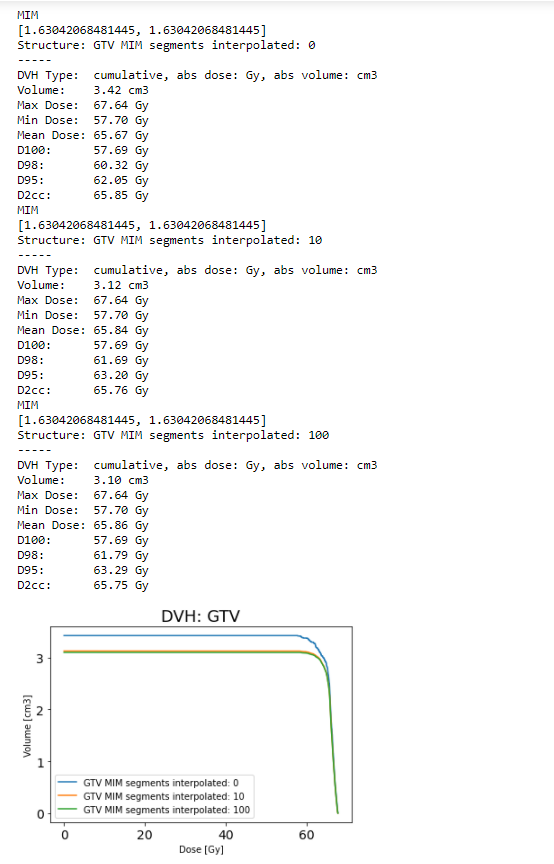
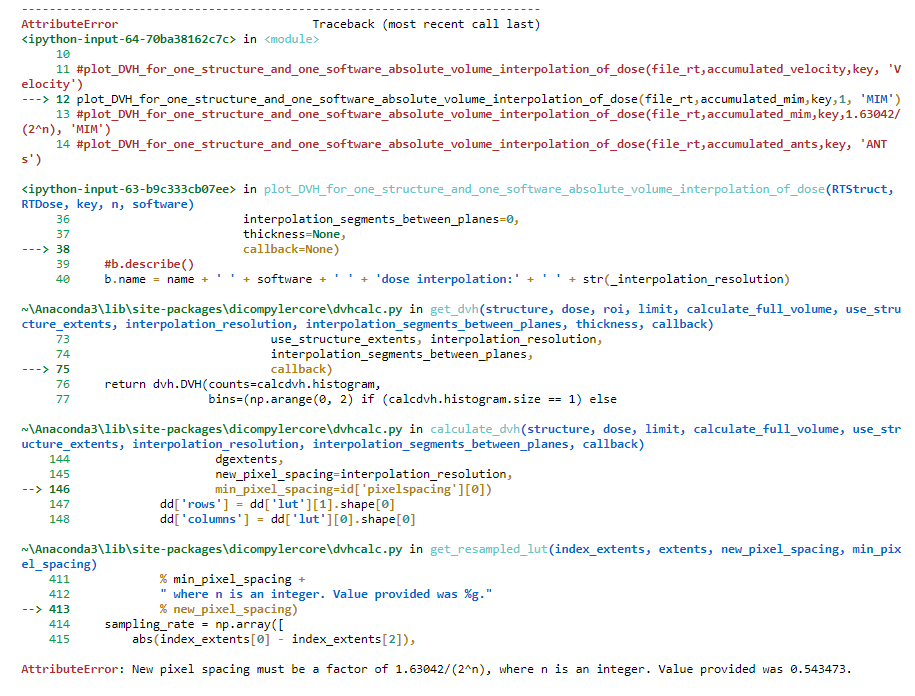
Isak Wahlstedt

Adit Panchal
On Jan 14, 2021, at 02:07, Isak Wahlstedt <isak.wa...@gmail.com> wrote:
Dear all,
A small addition to my previous post; it seems like I provided an incorrect input value of 0.543473 for the interpolation_resolution. I just corrected this and got the same error message (see below).
To view this discussion on the web visit https://groups.google.com/d/msgid/dicompyler/75fa5b8b-9637-4754-971a-c6a188821dedn%40googlegroups.com.
Dan Cutright
if dvh.volume < self.dvh_small_volume_threshold:
try:
kwargs["interpolation_resolution"] = (
self.rt_data["dose"].PixelSpacing[0]
/ self.dvh_high_resolution_factor,
self.rt_data["dose"].PixelSpacing[1]
/ self.dvh_high_resolution_factor,
)
if dicompylercore_version
== "0.5.5":
kwargs["interpolation_resolution"] = kwargs[
"interpolation_resolution"
][0]
kwargs[
"interpolation_segments_between_planes"
] = self.dvh_high_resolution_segments_between
try:
dvh_new =
dvhcalc.get_dvh(**kwargs)
except MemoryError:
kwargs["memmap_rtdose"]
= True
# dicompyler-core needs to re-parse the dose file
dvh_new = dvhcalc.get_dvh(**kwargs)
dvh = dvh_new
except
Exception as
e:
msg = (
"Small volume calculation failed, "
"using default
calculation"
)
push_to_log(e, msg=msg)
Isak Wahlstedt