Missing 'TrimmedSequence' column
105 views
Skip to first unread message
David Tork
Feb 17, 2023, 5:23:07 PM2/17/23
to dartR
Hi all,
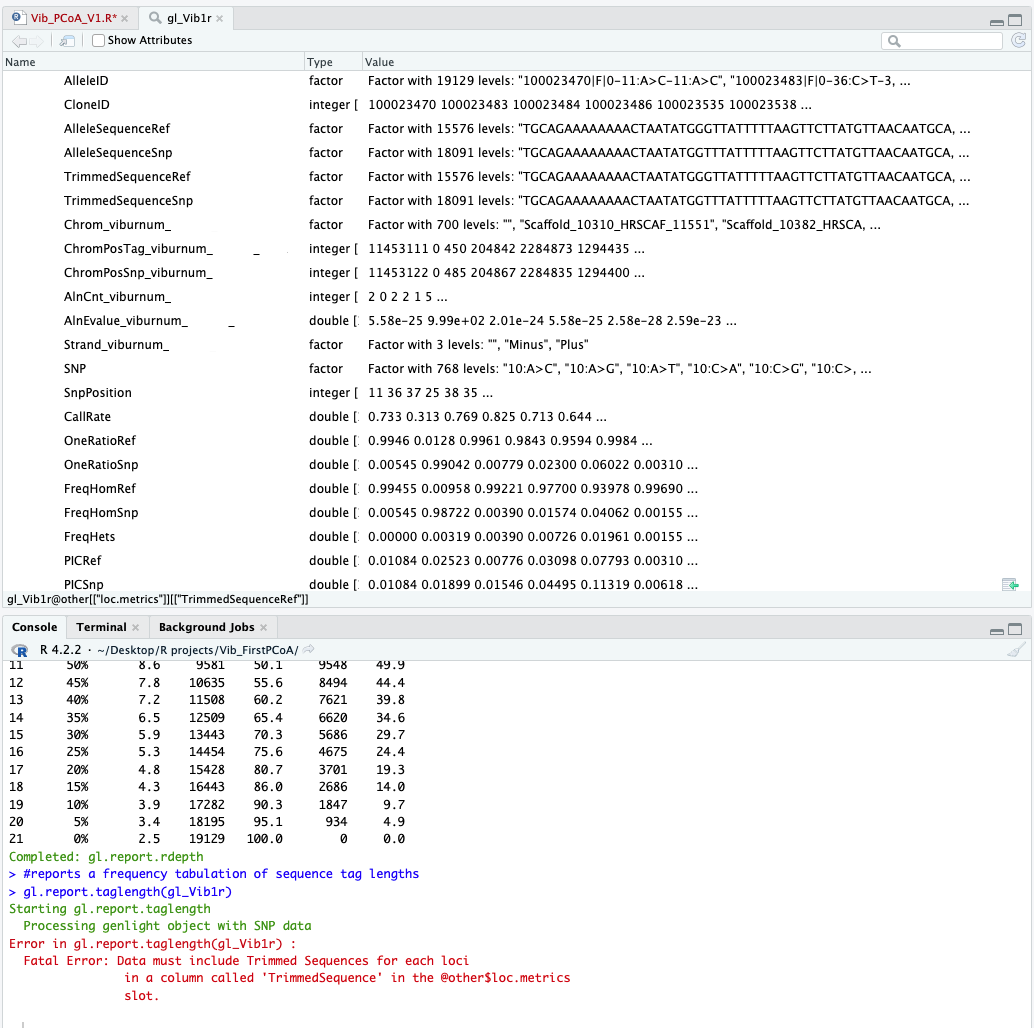
I am having trouble with the following functions:
gl.report.hamming()
gl.report.overshoot()
gl.report.taglength()
All return the same fatal error explaining that the 'TrimmedSequence' column is missing (see attached screenshot). Looking at the genlight object in @other$loc.metrics, you can see that there are two factors, 'TrimmedSequenceRef' and 'TrimmedSequenceSnp'. I'm guessing that one of these could simply be renamed to 'TrimmedSequence' and it would fix the error, but then which one? They differ in the number of levels (~18k vs ~15k).
Any insight would be appreciated.
Thanks,
David

Jose Luis Mijangos
Feb 22, 2023, 12:39:05 AM2/22/23
to dartR
Hi David,
We are working with DArT to determine a way to consistently use the same name for the trimmed sequence.
In the meantime, I would recommend creating the field "TrimmedSequence” using the “trimmedSequenceSNP” as shown below:
> your_gl$other$loc.metrics$TrimmedSequence <- your_gl$other$loc.metrics$TrimmedSequenceSnp
Cheers
Luis
David Tork
Mar 14, 2023, 12:27:31 PM3/14/23
to dartR
Hello Luis,


First, thank you for this suggestion. I was able to implement your workaround which allowed me to calculate the reporting functions mentioned previously.
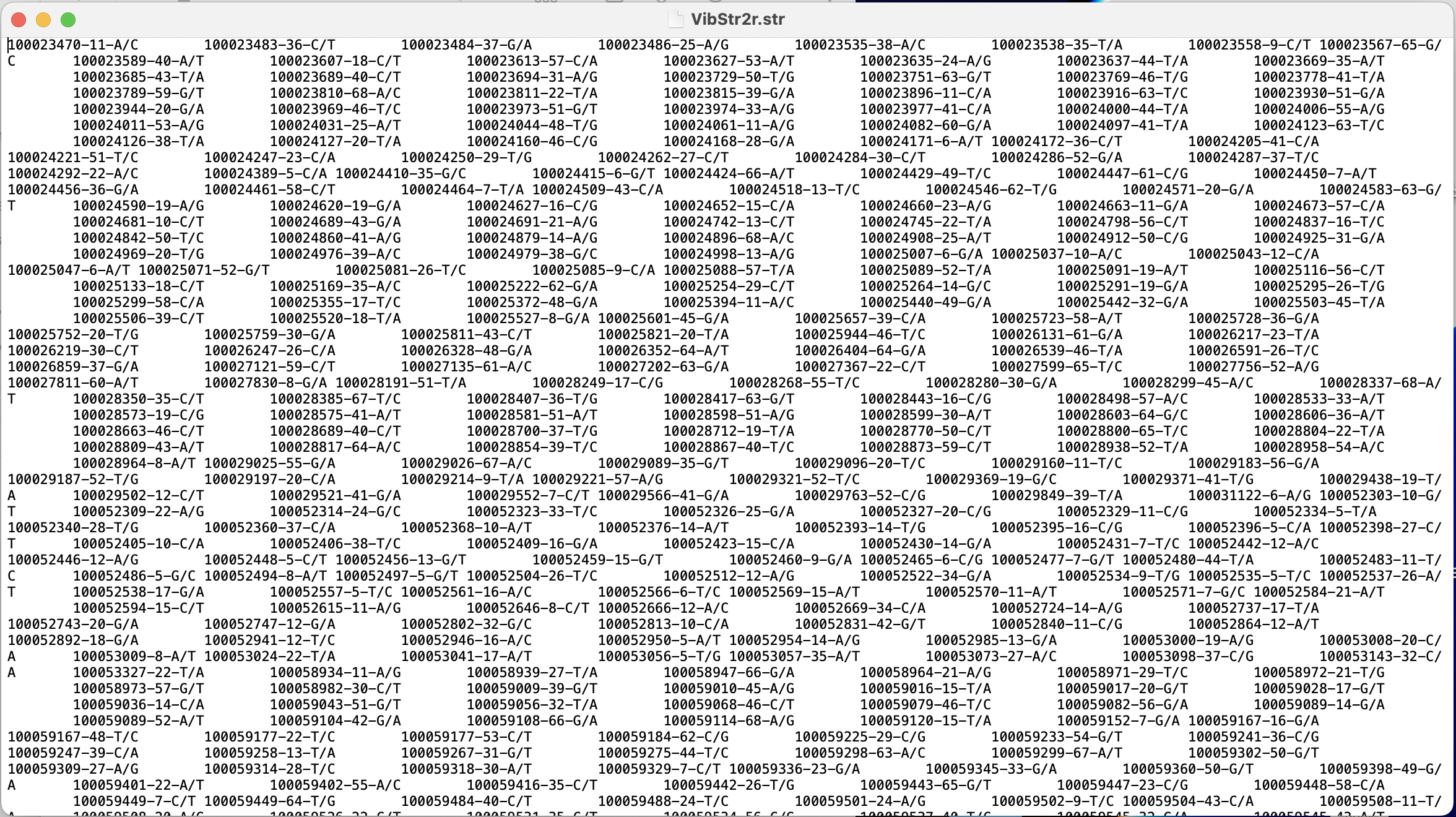
I have encountered another issue with the function gl2structure() which I am attaching to this thread, as I am wondering if it is also related to the formatting issues in my file.
The function works as expected and successfully outputs a .str file, however, for each individual in the dataset it creates two entries. So, for my 995 individuals, I end up with a structure file with 1990 rows. When I look at the file in a plaintext editor, I can confirm that each IndName appears twice. This occurs whether I use the 1-row or 2-row datasets as input. I also tested it on filtered and unfiltered datasets and obtained the same result.
I am using the default arguments as follows:
gl2structure(VibStr2r, outfile = "VibStr2r.str", outpath = "/R projects/Vib_Structure/")
Additionally, I spoke with a colleague who is also working with DartR & structure, and they indicated that the header rows on my structure file look different. Apparently theirs do not include so many spaces (screenshot below)

In the below screenshots, you can see a visual of what I mean about the duplicate IndName entries


Please let me know if you have any suggestions as I am unsure how to proceed.
Thanks,
David
David Tork
Mar 15, 2023, 10:38:24 AM3/15/23
to da...@googlegroups.com
Hi Luis,
Thank you for clarifying and pointing me to the pertinent information. My mistake for missing this very obvious portion of the documentation.
David
--
You received this message because you are subscribed to a topic in the Google Groups "dartR" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/dartr/plzh7giXTac/unsubscribe.
To unsubscribe from this group and all its topics, send an email to dartr+un...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/dartr/ca642b35-53fc-466c-8aa6-1df45013e23dn%40googlegroups.com.
Researcher
M.S. Plant Breeding and Molecular GeneticsDept. of Horticultural Science, University of Minnesota
Reply all
Reply to author
Forward
0 new messages
