Problems calculating mean() of population
223 views
Skip to first unread message
Ingo Miller
May 31, 2022, 1:27:01 AM5/31/22
to ctmm R user group
Hi Chris,
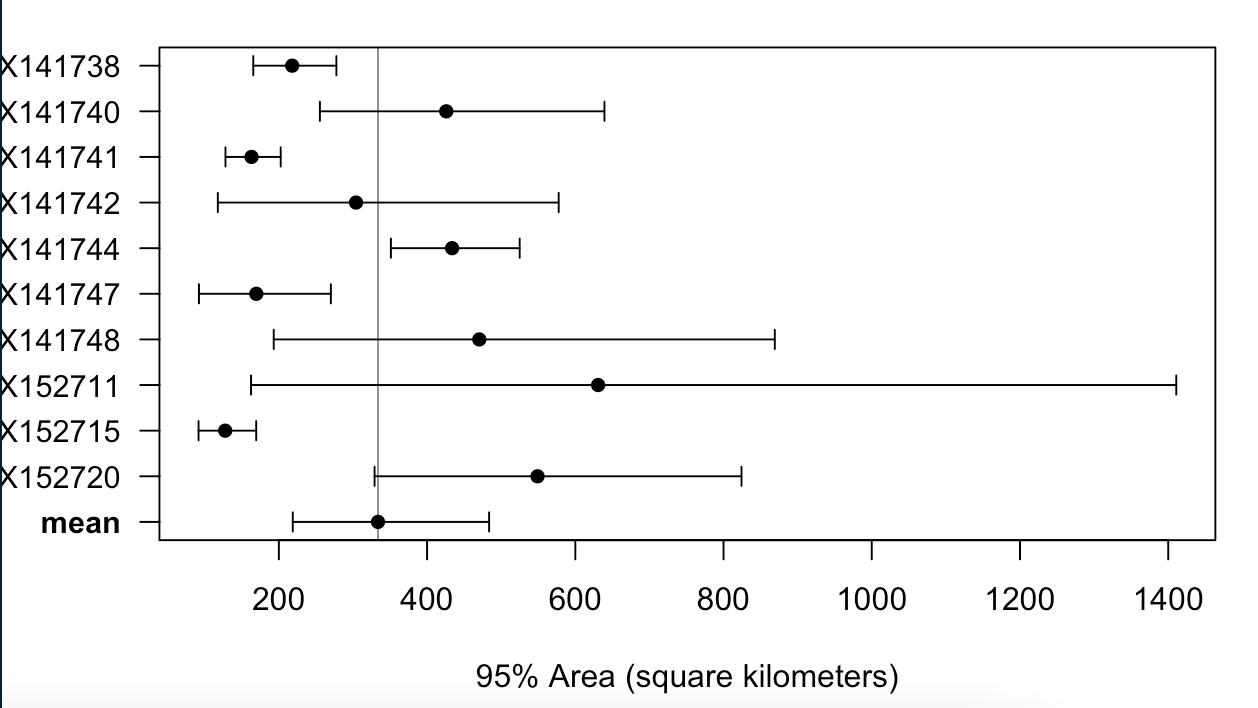
I want to calculate mean AKDEs for two subpopulations of sea turtles using the mean() functions. It works fine for one of the group of 5 turtles, however running mean() on the UDs of the other group of 10 individuals results in some weird error which I cannot determine what it means. The error is as following:
> mean(UDS_CI)
Error in SIGMA[i, P] <- matrix(SIGMA[i, "major", "major"], 2, 2) :
incorrect number of subscripts on matrix
Error in SIGMA[i, P] <- matrix(SIGMA[i, "major", "major"], 2, 2) :
incorrect number of subscripts on matrix
The same error occurs when I run mean() on the FIT object.
Do you know what the issue could be here?
The meta() function, on the other hand works resulting in the following:
> meta(UDS_CI)
ΔAICc
inverse-Gaussian 0.00000
Dirac-δ 53.31221
low est high
mean (km²) 218.77416726 333.7738223 483.6887881
CoV² (RVAR) 0.05705984 0.2975987 0.7328652
CoV (RSTD) 0.24926673 0.5692651 0.8933284
ΔAICc
inverse-Gaussian 0.00000
Dirac-δ 53.31221
low est high
mean (km²) 218.77416726 333.7738223 483.6887881
CoV² (RVAR) 0.05705984 0.2975987 0.7328652
CoV (RSTD) 0.24926673 0.5692651 0.8933284

Here's what I coded before to get there:
#Fitting error with uncalibrated data under a prior:
uere(CI.trj) <- NULL
# assign 10m error
uere(CI.trj) <- c(10)
# the default uncertainty is none for numerical assignments
UERE <- uere(CI.trj)
summary(UERE)
UERE$DOF
UERE$DOF[] <- 2
summary(UERE)
# assign the prior to the data
uere(CI.trj) <- UERE
fitting_function <- function(i)
{
T.GUESS <- ctmm.guess(CI.trj[[i]], CTMM = ctmm(error = T), interactive=F)
ctmm.select(CI.trj[[i]], T.GUESS, verbose = F, trace=2)
}
for(i in 1:length(CI.trj)) { T.FITS[[i]] <- fitting_function(i) }
#parallelizing function to speed things up
cl <- parallel::makeForkCluster(2)
doParallel::registerDoParallel(cl)
foreach(i = 1:3, .combine = 'c') %dopar% {
sqrt(i)
}
T.FITS <- foreach(i=1: length(CI.trj), .packages='ctmm') %dopar% { fitting_function(i) }
uere(CI.trj) <- NULL
# assign 10m error
uere(CI.trj) <- c(10)
# the default uncertainty is none for numerical assignments
UERE <- uere(CI.trj)
summary(UERE)
UERE$DOF
UERE$DOF[] <- 2
summary(UERE)
# assign the prior to the data
uere(CI.trj) <- UERE
fitting_function <- function(i)
{
T.GUESS <- ctmm.guess(CI.trj[[i]], CTMM = ctmm(error = T), interactive=F)
ctmm.select(CI.trj[[i]], T.GUESS, verbose = F, trace=2)
}
for(i in 1:length(CI.trj)) { T.FITS[[i]] <- fitting_function(i) }
#parallelizing function to speed things up
cl <- parallel::makeForkCluster(2)
doParallel::registerDoParallel(cl)
foreach(i = 1:3, .combine = 'c') %dopar% {
sqrt(i)
}
T.FITS <- foreach(i=1: length(CI.trj), .packages='ctmm') %dopar% { fitting_function(i) }
akde_function <- function(i)
{
print(i)
akde(CI.trj[[i]], T.FITS[[i]], weights=T, trace=T, grid=list(dr=500, align.to.origin=T))
}
UDS_CI <- foreach(i=1: length(CI.trj), .packages='ctmm') %dopar% { akde_function(i) }
{
print(i)
akde(CI.trj[[i]], T.FITS[[i]], weights=T, trace=T, grid=list(dr=500, align.to.origin=T))
}
UDS_CI <- foreach(i=1: length(CI.trj), .packages='ctmm') %dopar% { akde_function(i) }
parallel::stopCluster(cl)
names(UDS_CI) <- names(CI.trj)
plot(CI.trj, UD=UDS_CI)
meta(UDS_CI) # -> works
mean(UDS_CI)
names(UDS_CI) <- names(CI.trj)
plot(CI.trj, UD=UDS_CI)
meta(UDS_CI) # -> works
mean(UDS_CI)
I attach the data and ctmm objects as well.
Cheers,
Ingo
Christen Fleming
May 31, 2022, 10:15:06 AM5/31/22
to ctmm R user group
Hi Ingo,
This bug is already fixed on GitHub if you update.
Best,
Chris
Ingo Miller
Jun 13, 2022, 5:28:44 AM6/13/22
to ctmm R user group
Hi Chris,
I even tried that but seems like I I should have restarted my system. Anyway now it works fine. Thank you!
I do have another ussie though. I'm running several individuals through a parallelized foreach loop and now get the following error:
fitting_function <- function(i)
{
T.GUESS <- ctmm.guess(turtle.trj[[i]], CTMM = ctmm(error = T), interactive=F)
ctmm.select(turtle.trj[[i]], T.GUESS, verbose = F, trace=2)
}
FITS <- foreach(i=1: length(turtle.trj), .packages='ctmm') %dopar% { fitting_function(i) }
{
T.GUESS <- ctmm.guess(turtle.trj[[i]], CTMM = ctmm(error = T), interactive=F)
ctmm.select(turtle.trj[[i]], T.GUESS, verbose = F, trace=2)
}
FITS <- foreach(i=1: length(turtle.trj), .packages='ctmm') %dopar% { fitting_function(i) }
akde_function <- function(i)
{
print(i)
akde(turtle.trj[[i]], FITS[[i]], weights=T, trace=3, fast=F, PC='direct')
{
print(i)
akde(turtle.trj[[i]], FITS[[i]], weights=T, trace=3, fast=F, PC='direct')
UDS <- foreach(i=1: length(turtle.trj), .packages='ctmm') %dopar% { akde_function(i) }
Error in { : task 40 failed - " Data not in single coordinate system."
I have run thus data set before with lapply and it worked fine but took ages (that was months ago though).
Do you know what the cause could be here? It works perfectly fine for a smaller subset of the data with less individuals. So I'm not sure why I get this error here as the data was prepared exactly the same way. Would appreciate some help.
Cheers,
IM
Christen Fleming
Jun 13, 2022, 11:34:20 AM6/13/22
to ctmm R user group
Hi Ingo,
If you have imported individuals from multiple files, then you will want to get them into the same projection before fitting models, such as like this:
projection(turtle.trj) <- median(turtle.trj)
Best,
Chris
Reply all
Reply to author
Forward
0 new messages
