Interpretation of variograms
Mark Thomas
Hi Chris,
I hope you are well. I am new to the CTMM package and have read and listened to information around whether data plotted on the variograms is okay for home-range analysis.
I have these weird patterns from my data.
It is movement data from a Vulture in Peru. The timestamps are variable in that the time between fixes ranges from 1min, 10min, 20min, or 1 hour. For CTMM models does the data need to be standardised (thin data to a point every 10mins or one hour?).
This particular species the tag records until 8pm.
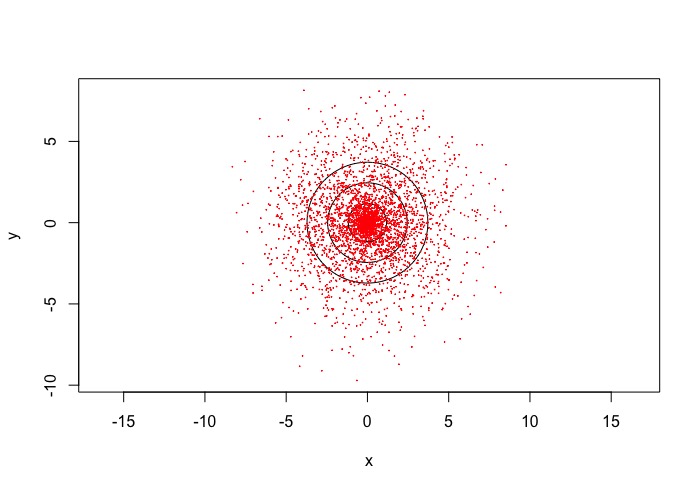
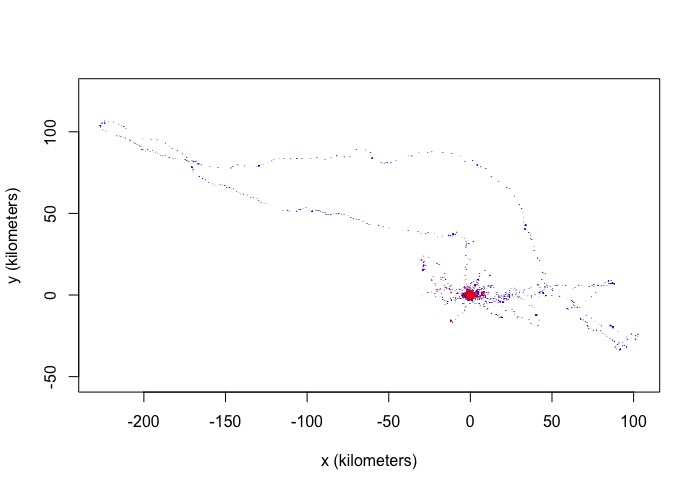
I looked at the colours of the tracks and the blue and red overlap, suggesting it is not migrating, however, there is a big loop in the data away from the concentrated area of points.
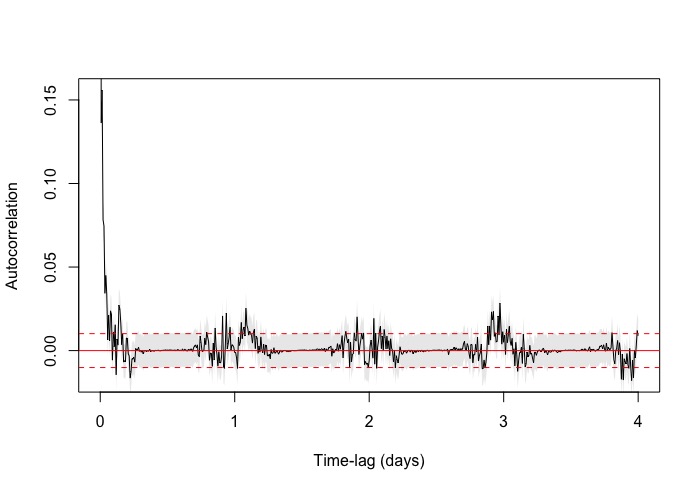
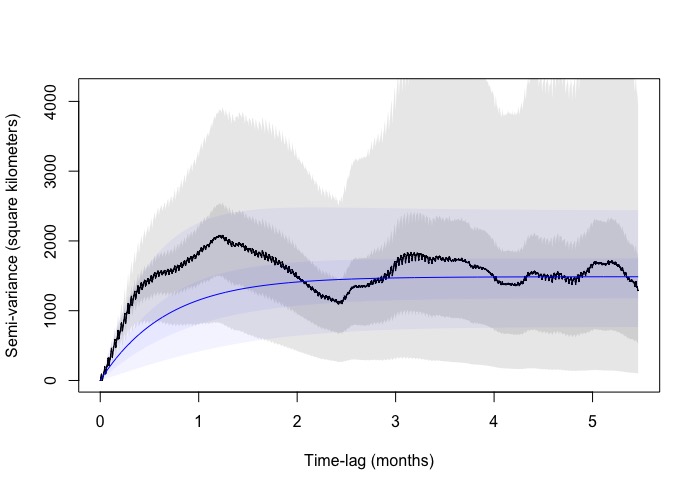
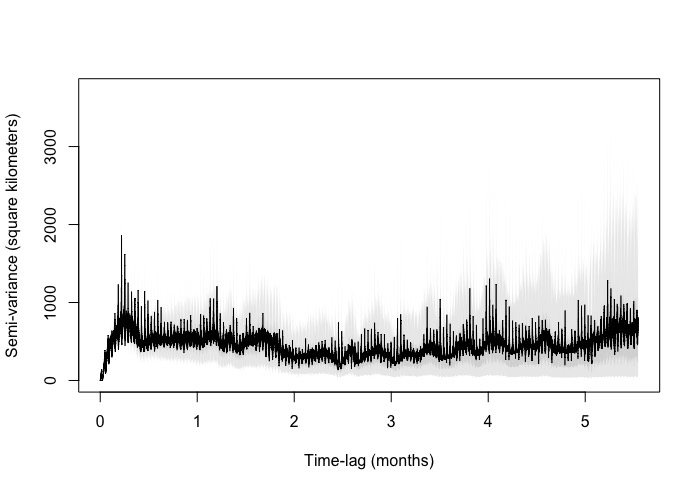
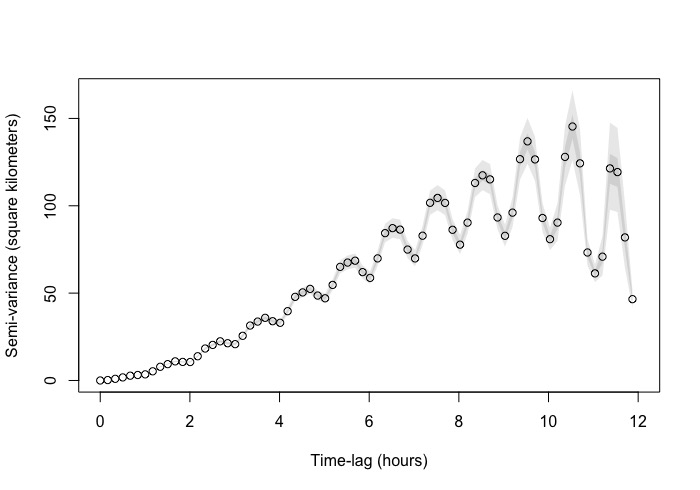
The zoomed in variogram increases linearly and then starts to do this zig zag pattern. I have a few other species where they do the same (some more extravagant) . The zoomed out version you can see the pattern more extensively. However, it does like it reaches asymptote albeit with the zig zag.
Are you able to advise if the variograms outputs are okay to model home-ranges? and if there's another check I can do within the ctmm workflow that helps me include / exclude individuals from HR analysis.
Many thanks in advance.
Christen Fleming
Mark Thomas
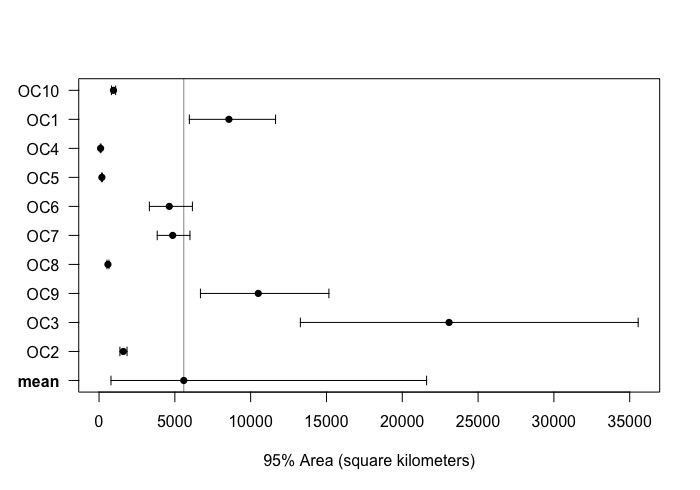
- For the models I ran:
FITS <- list()
for(i in 1:length(OCT))
{
# save the default guess value for each home range
GUESS <- ctmm.guess(OCT[[i]],interactive=FALSE)
# use the guess values to find the best model and save into list
FITS[[i]] <- ctmm.fit(OCT[[i]],GUESS,trace=3)
}
- And then (for each model):
- However, OC2, OC3, and OC4 I used:
- Because I kept getting this error:
Error: vector memory exhausted (limit reached?)
- When running without 'fast = FALSE'.
- When I thinned the data to 1 hour for OC3 it actually worked with fast=TRUE
Christen Fleming
- You should make sure the timestamps are importing correctly and the data are correct. GPS doesn't have millisecond accuracy.
- If keeping these tiny timesteps, the model fits are not likely to be any good without an error model, because this animal will not be displacing any appreciable distance within the timespan of 1 millisecond. With the error model included, the model fits should improve, but the default arguments of weighted akde() will still have some trouble, as a 1 millisecond grid is too large for the "fast" algorithm and you run out of memory. You either need to increase the dt argument to something like 1 minute or switch to the non-gridded fast=FALSE,PC='direct' option (if your datasets aren't too large for that algorithm to be prohibitive).
Mark Thomas