data import - UTM?
150 views
Skip to first unread message
Erin Netoskie
Feb 24, 2019, 5:13:58 PM2/24/19
to ctmm R user group
Aloha,
I am trying to learn how to use the ctmm package for my thesis by creating the kernel densities for home range analysis. My data was collected using automated telemetry towers, but I am located in Hawaii where we use UTM rather than lat long. The data does not belong to me and I have been told not to upload it to Movebank as it will be made public later. I am having issues getting the data correctly formatted so it will import correctly. My column headers are band.num (bird ID ex: 93229899), date (1/17/18), time (11:30 - using military time), UTM East (251402.412), and UTM North (2174029.523).
I think part of the reason I am getting so many errors is the UTM and date and time formats, but everything I try with as.POSIXct and as.telemetry is not working. Do you have any advice?
Thank you,
Erin
Christen Fleming
Feb 26, 2019, 10:44:48 AM2/26/19
to ctmm R user group
Aloha Erin,
I've just put support for importing UTM coordinates in the development branch, which you can install via devtools::install_github("ctmm-initiative/ctmm") . Otherwise you can inverse project from UTM to long-lat with rgdal::project().
You will need a column designating the UTM zone.
You will need to rename band.num to a Movebank attribute like animal.ID or individual.local.identifier. I can add support for band.num in the future.
You will need to paste() the date and time together into one timestamp column. You might also need to provide a time format argument if as.POSIXct() can not automatically figure out your timestamp format.
Best,
Chris
Christen Fleming
Feb 26, 2019, 11:33:19 AM2/26/19
to ctmm...@googlegroups.com
I slipped a bug into ctmm.select() on Linux & MacOS. So, unless you are running Windows, please wait until the update tonight before attempting to run the updated package with UTM support.
EDIT: Fixed on GitHub now.
Best,
Chris
Erin Netoskie
Feb 28, 2019, 8:34:55 PM2/28/19
to ctmm R user group
Thank you so much for advice and quick response!
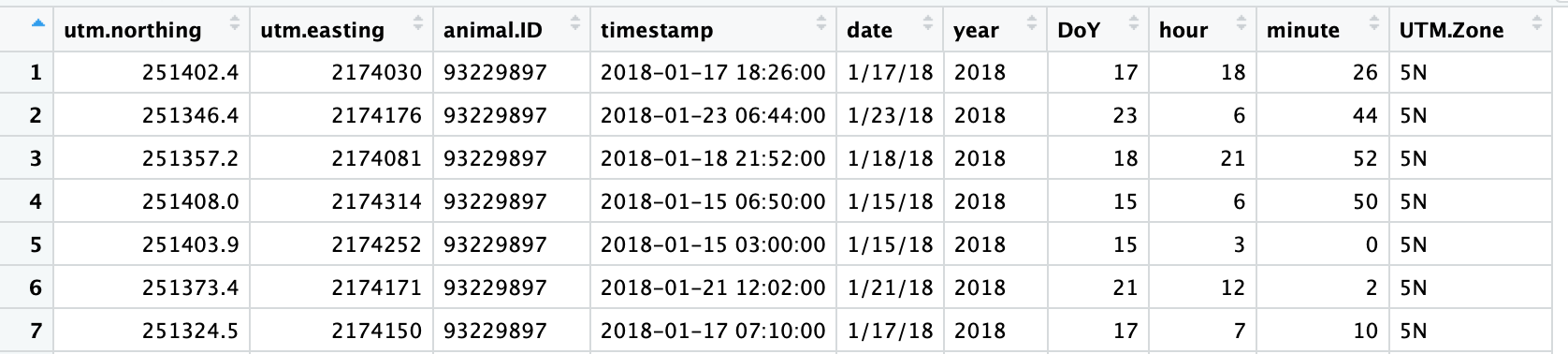
I am now getting a different error about undefined columns (I apologize if these are simple questions, I am still new to R).
I believe I used as.POSIXct() correctly - I've attached a screenshot of what my data looks like.

This is my as.telemetry code:
hr4.dat <- as.telemetry(ctmm4.dat,timeformat = "%Y-%m-%d %H:%M:%S",timezone="Pacific/Honolulu",timeout=Inf,na.rm="row")
Do you have any advise for this error?
Mahalo,
Erin
Christen Fleming
Mar 1, 2019, 3:07:11 PM3/1/19
to ctmm R user group
Hi Erin,
Do you mind sending me a sample of the data that can reproduce this error, either via message or email? The data and timestamp format look okay to me if you updated the ctmm package from GitHub recently.
Best,
Chris
Erin Netoskie
Mar 27, 2019, 4:10:14 PM3/27/19
to ctmm R user group
Hi Chris,
Is there a way in ctmm to calculate the centroid of the akde outputs? I want to know what the center of their home range is and then calculate the distance of each telemetry point from the center. Or is there another package you would recommend for this?
Thanks,
Erin
Christen Fleming
Mar 28, 2019, 8:44:12 PM3/28/19
to ctmm R user group
Hi Erin,
The mean location is already estimated in the fit object: FIT$mu
The mode location you can find from the UD object either at the maximum of UD$PDF or (equivalently) at the minimum of UD$CDF. These are both 2D arrays. The grid locations are stored in UD$r$x and UD$r$y.
Best,
Chris
Reply all
Reply to author
Forward
0 new messages
