R's apparent inability to handle UTF-8 encoding when cleaning up corpus files in R and outputting them into .txt
Juho Kristian Ruohonen
I have a manually compiled 1.5-million-word corpus of online newswriting -- about 3,000 articles, each stored in one textfile. Now I've been trying to concatenate these files into one single file (in order to have the corpus in a more easily copy-pasteable form for the free online CLAWS5 tagger). But the corpus needs to be cleaned up first in order for the tagger to work properly. Typographic apostrophes (’‘) and quotation marks (”“) must be replaced with regular ones and also separated from words wherever they don't indicate a missing letter, etc. However, I am finding that whenever I either
1.perform any pattern replacement on the corpus vector with gsub or
2.add anything other than the vector itself and the sep="\n" argument to the vector when catting it to an output file,
the output file becomes tainted with myriad junk characters such as � etc, and not only where the "exotic" characters are, but also at the beginning of each appended vector. The junk only appears if I perform gsub replacements before outputting or add extra input with cat when outputting the vectors back out. If I simply scan the textfiles and then cat them back out using sep="\n", they stay clean. But then I cannot do the cleanup. Both Word and OpenOffice Writer badly choke when doing search and replace on a textfile this large, and they don't even support backreferences, so they are no substitute for R.
Well, I have identified the problem -- it is the UTF-8 encoding of the textfiles. This problem is entirely absent in another corpus of mine whose textfiles are in ANSI encoding. However, I've learned the hard way that AntConc cannot display that encoding properly. Even if it somehow could, however, manually changing the encoding of all 2,887 textfiles constituting the corpus would take all day. And before you say don't use AntConc for concordancing when you can use R, I tried that too. My regex is very long and complex -- about 1.5 pages of continuous regex in font size 12. Strapply just malfunctions with it (and yes, I did change the backslashes into double backslashes for R).
Executive summary: How can I make R handle UTF-8-encoded textfiles without character encoding hell? Would changing my Windows locale to English -- American help? That would be a hassle since I do need to type in my own language (which uses [äöå]), but would it even help?
Matías Guzmán Naranjo
--
You received this message because you are subscribed to the Google Groups "CorpLing with R" group.
To unsubscribe from this group and stop receiving emails from it, send an email to corpling-with-r+unsubscribe@googlegroups.com.
To post to this group, send email to corpling-with-r@googlegroups.com.
Visit this group at https://groups.google.com/group/corpling-with-r.
For more options, visit https://groups.google.com/d/optout.
Juho Kristian Ruohonen
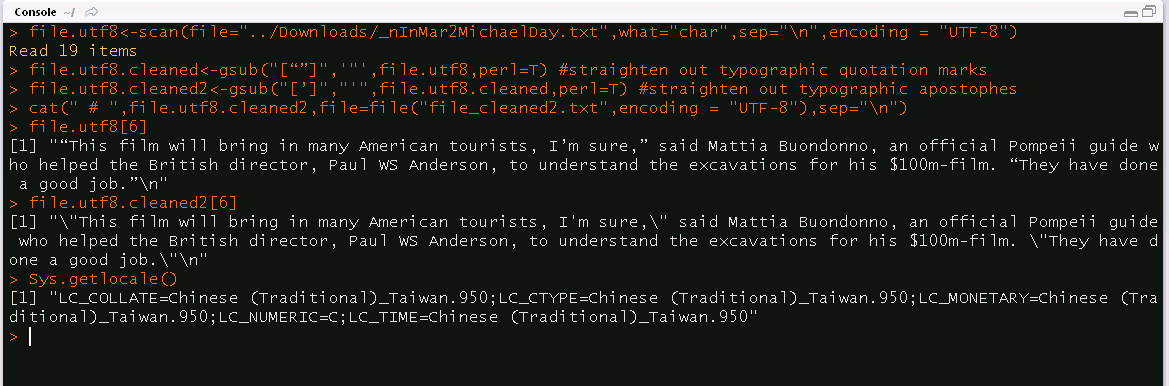
file1<-scan(file="_nInMar2MichaelDay.txt",what="char",sep="\n")
file.cleaned<-gsub("[”“]",'"',file1,perl=T) #straighten out typographic quotation marks
file.cleaned2<-gsub("[’‘]","'",file.cleaned,perl=T) #straighten out typographic apostophes
cat(" # ",file.cleaned2,file="file_cleaned2.txt",sep="\n")The substitutions either do nothing or introduce junk characters alongside the replacement.
On Stefan's Linux system, reportedly, everything works fine and displays correctly.
Here's something absolutely key: when I scan this utf-8 textfile into an R vector, those typographic characters turn into junk characters in R. This is why some of the substitutions simply do nothing.
Here's the next thing I tried:
file.utf8<-scan(file="_nInMar2MichaelDay.txt",what="char",sep="\n",encoding = "UTF-8")
file.utf8.cleaned<-gsub("[“”]",'"',file.utf8,perl=T) #straighten out typographic quotation marks
file.utf8.cleaned2<-gsub("[’]","'",file.utf8.cleaned,perl=T) #straighten out typographic apostophes
cat(" # ",file.utf8.cleaned2,file="file_cleaned2.txt",sep="\n")Result: Now the special characters display correctly in R, but the substitutions do nothing. Gsub fails to even recognize those characters (even if I copypaste them directly from the R's printout), so nothing gets replaced.
I suppose the problem must be either with the Windows version of R (mine is 3.2.4) or with my non-English locale. ://
To unsubscribe from this group and stop receiving emails from it, send an email to corpling-with...@googlegroups.com.
To post to this group, send email to corplin...@googlegroups.com.
Alex Perrone

<_nInMar2MichaelDay.txt>
Juho Kristian Ruohonen
This is what I tried: I went to Control Panel and changed Formats, Location, Default Input Language, and System Locale to English (United States), United States, English (United States), and English (United States), respectively. I've already set default encoding to UTF-8 in RStudio's Tools -> Global Options -> Code -> Saving. And Sys.getlocale() prints the following:
"LC_COLLATE=English_United States.1252;LC_CTYPE=English_United States.1252;LC_MONETARY=English_United States.1252;LC_NUMERIC=C;LC_TIME=English_United States.1252"
The problem still remains, however. The UTF-8 textfile still displays smart quotes and apostrophes as garbage within R, and gsub attempts either do nothing or result in more garbage. The last thing I tried was the following:
[1] ""
Warning message: In Sys.setlocale("LC_ALL", "en_US.UTF-8") : OS reports request to set locale to "en_US.UTF-8" cannot be honored
Not sure what to try next. Anyone know a convenient way to change the encoding of 2887 textfiles in one go?
Matías Guzmán Naranjo
To unsubscribe from this group and stop receiving emails from it, send an email to corpling-with-r+unsubscribe@googlegroups.com.
To post to this group, send email to corpling-with-r@googlegroups.com.
Alex Perrone
Alvin Chen
Alvin C.-H. Chen, Ph.D. (陳正賢)
Assistant Professor
Department of English
National Taiwan Normal University
國立台灣師範大學英語系
To unsubscribe from this group and stop receiving emails from it, send an email to corpling-with-r+unsubscribe@googlegroups.com.
To post to this group, send email to corpling-with-r@googlegroups.com.
Hardie, Andrew
All this sounds very much like the problem is with the character encoding of the script. That is, if the script is coded in Win 1252 (which most Windows editors will generate by default), doing replacements of literal quote marks will not produce the correct results (since the literals are represented as single bytes in the range x80 to x9f in 1252, which will match partial UTF-8 characters, thus scrambling the content). The script and the target files need to have the same encoding for replacements of this kind to work.
I suggest examining the script in an editor that makes the encoding transparent (e.g. Notepad++) and converting to UTF-8 if necessary…
best
Andrew.
Alvin Chen
To unsubscribe from this group and stop receiving emails from it, send an email to corpling-with-r+unsubscribe@googlegroups.com.
To post to this group, send email to corpling-with-r@googlegroups.com.
Visit this group at https://groups.google.com/group/corpling-with-r.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to the Google Groups "CorpLing with R" group.
To unsubscribe from this group and stop receiving emails from it, send an email to corpling-with-r+unsubscribe@googlegroups.com.
To post to this group, send email to corpling-with-r@googlegroups.com.
Visit this group at https://groups.google.com/group/corpling-with-r.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to the Google Groups "CorpLing with R" group.
To unsubscribe from this group and stop receiving emails from it, send an email to corpling-with-r+unsubscribe@googlegroups.com.
To post to this group, send email to corpling-with-r@googlegroups.com.
Hardie, Andrew
Yeah, nearly all Windows text editors insert the BOM in UTF-8 by default; this is generally not what you want esp., if you’re going to be concatenating stuff, as you then get spurious zero-width-no-break-spaces mid file. When reading UTF-8 files, unless they are from a known-safe source, I normally check for and discard the BOM first of all. (Which is a pretty simple regex: “^\x{feff}” or “^\xef\xbb\xbf” as bytes.)
Andrew.
From: corplin...@googlegroups.com [mailto:corplin...@googlegroups.com]
On Behalf Of Alvin Chen
Sent: 18 December 2016 23:41
To: corplin...@googlegroups.com
Subject: Re: [CorpLing with R] R's apparent inability to handle UTF-8 encoding when cleaning up corpus files in R and outputting them into .txt
Another thing.
In my output text file, while all the curly quotes are taken care of (converted successfully), there is a <U+FEFF> at the beginning of the text file. (p.s. The output is in UTF-8) Is it because the original file from Juho is UTF-8-BOM? But i can't scan() the input file with that encoding successfully though. Processing UTF-8 files in windows is really discouraging.
Alvin
On Mon, Dec 19, 2016 at 7:01 AM, Hardie, Andrew <a.ha...@lancaster.ac.uk> wrote:
All this sounds very much like the problem is with the character encoding of the script. That is, if the script is coded in Win 1252 (which most Windows editors will generate by default), doing replacements of literal quote marks will not produce the correct results (since the literals are represented as single bytes in the range x80 to x9f in 1252, which will match partial UTF-8 characters, thus scrambling the content). The script and the target files need to have the same encoding for replacements of this kind to work.
I suggest examining the script in an editor that makes the encoding transparent (e.g. Notepad++) and converting to UTF-8 if necessary…
best
Andrew.
From:
corplin...@googlegroups.com [mailto:corplin...@googlegroups.com]
On Behalf Of Alvin Chen
Sent: 18 December 2016 22:38
To: corplin...@googlegroups.com
Subject: Re: [CorpLing with R] R's apparent inability to handle UTF-8 encoding when cleaning up corpus files in R and outputting them into .txt
Hi,
Non-English locale users here as well. Here's my print-screen.

But something weird, though. At first, I tried Juho's script and I ran into the same problem. What I did was that I printed the lines with the curly quotes in console and then copied those curly quotes (from the console) back to the regular expressions in the script. And after that, everything works fine. Still no idea why....
Alvin
*****
Alvin C.-H. Chen, Ph.D. (陳正賢)
Assistant Professor
Department of English
National Taiwan Normal University
國立台灣師範大學英語系
Taipei City, 106, Taiwan
Alex Perrone
"utf-8-sig") for its Notepad program: Before any of the Unicode characters
is written to the file, a UTF-8 encoded BOM (which looks like this as a byte
sequence: 0xef, 0xbb, 0xbf) is written."On Dec 18, 2016, at 6:53 PM, Hardie, Andrew <a.ha...@lancaster.ac.uk> wrote:
Yeah, nearly all Windows text editors insert the BOM in UTF-8 by default; this is generally not what you want esp., if you’re going to be concatenating stuff, as you then get spurious zero-width-no-break-spaces mid file. When reading UTF-8 files, unless they are from a known-safe source, I normally check for and discard the BOM first of all. (Which is a pretty simple regex: “^\x{feff}” or “^\xef\xbb\xbf” as bytes.)Andrew.From: corplin...@googlegroups.com [mailto:corplin...@googlegroups.com] On Behalf Of Alvin Chen
Sent: 18 December 2016 23:41
To: corplin...@googlegroups.com
Subject: Re: [CorpLing with R] R's apparent inability to handle UTF-8 encoding when cleaning up corpus files in R and outputting them into .txt
Another thing.In my output text file, while all the curly quotes are taken care of (converted successfully), there is a <U+FEFF> at the beginning of the text file. (p.s. The output is in UTF-8) Is it because the original file from Juho is UTF-8-BOM? But i can't scan() the input file with that encoding successfully though. Processing UTF-8 files in windows is really discouraging.
Alvin
On Mon, Dec 19, 2016 at 7:01 AM, Hardie, Andrew <a.ha...@lancaster.ac.uk> wrote:
All this sounds very much like the problem is with the character encoding of the script. That is, if the script is coded in Win 1252 (which most Windows editors will generate by default), doing replacements of literal quote marks will not produce the correct results (since the literals are represented as single bytes in the range x80 to x9f in 1252, which will match partial UTF-8 characters, thus scrambling the content). The script and the target files need to have the same encoding for replacements of this kind to work.I suggest examining the script in an editor that makes the encoding transparent (e.g. Notepad++) and converting to UTF-8 if necessary…bestAndrew.From: corplin...@googlegroups.com [mailto:corplin...@googlegroups.com] On Behalf Of Alvin Chen
Sent: 18 December 2016 22:38
To: corplin...@googlegroups.com
Subject: Re: [CorpLing with R] R's apparent inability to handle UTF-8 encoding when cleaning up corpus files in R and outputting them into .txt
Hi,Non-English locale users here as well. Here's my print-screen.
<image001.png>
the output file becomes tainted with myriad junk characters such as � etc, and not only where the "exotic" characters are, but also at the beginning of each appended vector. The junk only appears if I perform gsub replacements before outputting or add extra input with cat when outputting the vectors back out. If I simply scan the textfiles and thencat them back out using sep="\n", they stay clean. But then I cannot do the cleanup. Both Word and OpenOffice Writer badly choke when doing search and replace on a textfile this large, and they don't even support backreferences, so they are no substitute for R.
Hardie, Andrew
The main point I was getting at was the encoding of the script, rather than the corpus text file. The issue of BOM in the input is a side point really (although it does, I think, explain the appearance of  in JKR’s “junk”-filled output). – AH.
Juho Kristian Ruohonen
All my code in RStudio (when pasted into Notepad++) is reportedly encoded in "UTF-8". This would be the problem according to Andrew's theory. However, RStudio does not seem to have the option of changing the encoding of one's code to "UTF-8-BOM". Even when I check "select all encodings", "UTF-8-BOM" (or even "UTF-8-SIG") does not figure on the list of options.
The next thing I tried was to scan the file with the encoding="UTF-8" argument so that it is displayed correctly in R, write my gsub commands in Notepad++ instead of R, encode those commands in UTF-8-BOM, paste them into the R console and execute. Nah, nothing gets substituted. Apparently R forces the input into UTF-8 as soon as it is introduced.
The last thing I tried was to scan the file as usual, with no encoding specified (which turns smart quotes and apostrophes into junk in my locale), then try to clean up the junk by manually pasting it from the R printout into the regex in the gsub call. This does not work either -- the replacement includes junk and/or is not what was specified.
As mentioned in an earlier post, I've already changed everything in Windows' Control Panel to "English -- United States". The R locale is reportedly
"LC_COLLATE=English_United States.1252;LC_CTYPE=English_United States.1252;LC_MONETARY=English_United States.1252;LC_NUMERIC=C;LC_TIME=English_United States.1252".
Trying to change it into anything that is specifically UTF-8 always returns
Warning message: In Sys.setlocale("LC_ALL", "en_US.UTF-8") : OS reports request to set locale to "en_US.UTF-8" cannot be honored
Some users in the link provided by Alex suggested editing the .Renviron file. No such file seems to exist anywhere on my hard drive.
Looks like a batch conversion of all 2,887 files into a less troublesome encoding than UTF-8-BOM is the next step.
Hardie, Andrew
>> All my code in RStudio (when pasted into Notepad++) is
If you copy paste, then the encoding will change to whatever N++ is currently set to. (To the best of my understanding, internally in Windows operations, everything is UTF-16LE, with translations at either end as needed.)
>> write my gsub commands in Notepad++ instead of R, encode those commands in UTF-8-BOM, paste them into the R console and execute
This won’t work for the same reason. The encoding will be adjusted by Windows upon paste into whatever encoding your R console is set for. And it seems pretty clear your console encoding is 1252 – witness all the 1252s in your locale string.
Anyway: My apologies for giving unhelpful advice based on a misunderstanding, I had thought you were running the script from a file whose encoding could be checked, rather than a pane in RStudio. Here’s some advice that might be more helpful, fingers crossed:
· I believe the reason that you get an error upon changing to “en_US.UTF-8” is that this is a Linux-style locale. “English_United States.65001” would be, I think, the Windows equivalent. (65001 = Windows-speak for UTF-8.) However, I have seen some notes online that say you can’t use any locale that includes 65001 – notably the MSDN documentation on setlocale() to be found here: https://msdn.microsoft.com/en-us/library/x99tb11d.aspx . So this may well also fail.
· If it fails, I can suggest two possibilities.
· One. Save your script in a file and invoke it with command-line R rather than from R studio. Then, you can make sure you have the correct encoding of the script by examining that file as it is on disk, using N++ or other tool, rather than trying to set the console encoding.
· Two, change the literal “,”,‘,’ etc. in your regexes to hexadecimal codes which will not be vulnerable to encoding problems.
>> RStudio does not seem to have the option of changing the encoding of one's code to "UTF-8-BOM". Even when I check "select all encodings", "UTF-8-BOM" (or even "UTF-8-SIG") does not figure on the list of options.
That’s because these are not generally recognised encodings. They are somebody’s attempt to treat the versions with/without BOM as two separate versions of Unicode, but an attempt that has not had wide adoption.
best
Andrew.
From: corplin...@googlegroups.com [mailto:corplin...@googlegroups.com]
On Behalf Of Juho Kristian Ruohonen
Sent: 19 December 2016 09:40
To: CorpLing with R
Subject: Re: [CorpLing with R] R's apparent inability to handle UTF-8 encoding when cleaning up corpus files in R and outputting them into .txt
According to Notepad++, the encoding of the corpus files (including the sample one) is "UTF-8-BOM". Hence the BOMs at the beginning of each file. But while annoying, the BOMs are only a minor annoyance compared to the mess created by smart quotes and apostrophes.
--
Juho Kristian Ruohonen
using hexadecimal codes for the characters in the gsub call did not fail. This is quite an ingenious workaround. It effectively solves my problem.
Also, Notepad++ apparently blows both Word and OpenOffice Writer right out of the water at handling huge textfiles. It is so efficient that I don't even need R for character replacements in a corpus this size (unless backreferences are required). I was previously unaware of this powerful tool.
Many thanks to everyone for their help, especially Andrew.
Stefan Th. Gries
Juho Kristian Ruohonen
file.utf8<-scan(file="n_e&s2may16.txt",what="char",sep="\n",encoding = "UTF-8") #Scan the file and specify the encoding as utf-8 so that it gets displayed correctly in R. The problem, of course, is that the file is not truly encoded in UTF-8 but UTF-8-BOM (courtesy of Notepad), so there is a mismatch of encodings between R and the scanned vector. Matching characters literally will therefore not work.
file.utf8.cleaned<-gsub("[\u201C\u201D]","\u0022",file.utf8,perl=T) #straighten out typographic quotation marks
file.utf8.cleaned2<-gsub("[\u2018\u2019]","\u0027",file.utf8.cleaned,perl=T) #straighten out typographic apostrophes
cat(" # ",file.utf8.cleaned2,file="file_utf8_cleaned2.txt",sep="\n") #output the file.A minor remaining difficulty is the BOM garbage that will appear at the start of every appended vector in the output file. I dunno how to fix that using gsub(), that's some next-level sh*t, as the BOM garbage both is and isn't there at the same time (it is absent when I print out the vector in RStudio or try to match it with strapply, but present in the preview screen, and it appears in the output when the vector is cat()ted to an external file.) But Notepad++ can handle those in a matter of seconds.
Stefan Th. Gries
Hardie, Andrew
Hmm, Juho, from your comments in this code snippet, I see I explained the issue poorly once again.
The problem is *not* that “the file is not truly encoded in UTF-8 but UTF-8-BOM”. BOM does cause the problems in concatenating, of course, but that’s a separate issue to the punctuation searching.
Rather, the problem is that your console (and, therefore, your code) is using Windows-1252, which mismatches your data files encoded as UTF-8.
Because your code is in 1252, when you include in your regex, say, a literal curly quote “ , its byte representation is 0x93 . But in your files, which are UTF-8, the same character (U+201C) has the byte representation 0xE2, 0x80, 0x9C. The problem is then that searching for 0x93 does not match 0xE2, 0x80, 0x9C!
However, searching for 0x93 may match one of the bytes of some other UTF-8 sequence(s). Once 0x93 is replaced, any such sequence is corrupted, and the overall string may not be recognisable to R and/or the OS as UTF-8 any longer (thus it is treated as an 8-bit encoding, with many many things rendering incorrectly).
Using a \u escape instead works because (to oversimplify a bit) it causes R to insert the correct UTF-8 byte sequence into the string, regardless of the encoding of the file or console that contains the code.
I hope this fuller explanation clarifies the issue.
>> I dunno how to fix that using gsub(), that's some next-level sh*t
Does search-and-replace on the regex ^\ufeff (with empty replacement string) not work to remove BOM? It ought to.
best
Andrew.
From: corplin...@googlegroups.com [mailto:corplin...@googlegroups.com]
On Behalf Of Juho Kristian Ruohonen
Sent: 20 December 2016 13:46
To: CorpLing with R
Subject: Re: [CorpLing with R] R's apparent inability to handle UTF-8 encoding when cleaning up corpus files in R and outputting them into .txt
Hmm. My bad. I tried different character codes until I found the ones that matched. I think they're actually Unicode character codes instead of hexadecimal (I didn't know the difference at the time). So here's what I did that works:
--
