Equilibrium steady-state and parameter estimation
61 views
Skip to first unread message
Deepa
Nov 1, 2022, 1:51:53 PM11/1/22
to COPASI User Forum
Dear All,

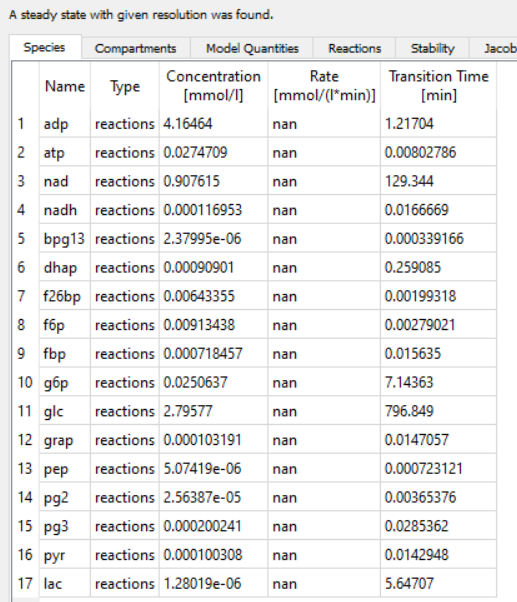

The following set of reactions are present in our model,

I ran the steady-state analysis task and I see the system reaches a steady state.

However, when I remove the ATP formation reaction from the model, the system reaches an equilibrium steady state where the fluxes are zero.

I am not sure if I can do a parameter estimation (using steady-state data) when the initial state of the model doesn't reach a steady state.
When I try to fix one of the nucleotides (nadh or nad) to constant, the system reverts to steady state. But if I fix both nadh and nad, the system attains equilibrium steady state again.
I'm trying to understand if I could make changes to the model so that the system doesn't reach an equilibrium steady state when the ATP formation reaction is removed.
Suggestions on this will be of great help.
Thank you very much for your time and kind attention,
Deepa
Reply all
Reply to author
Forward
0 new messages
