Error in the mgPipe tutorial
32 views
Skip to first unread message
Una Wu
Mar 1, 2023, 1:15:32 AM3/1/23
to COBRA Toolbox
Hello alls,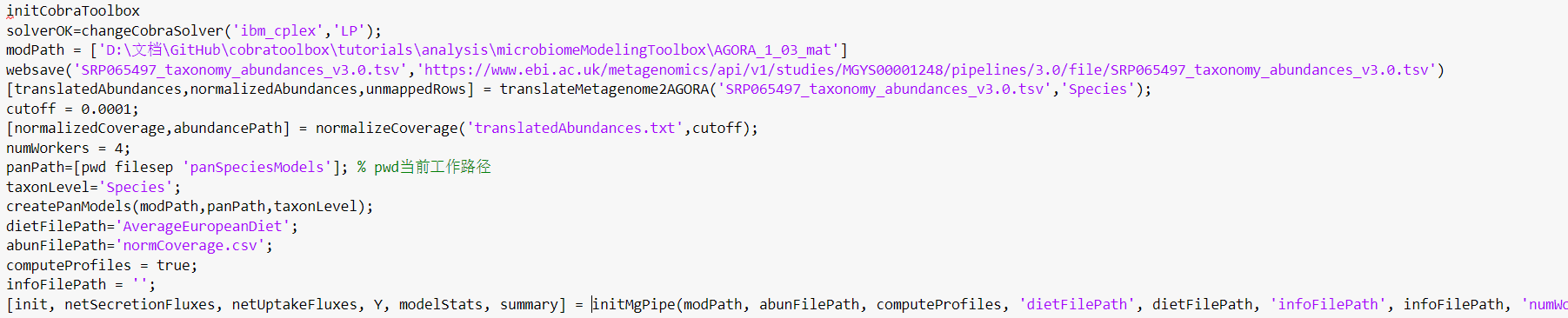
I run the tutorial_mgPipe.mlx file successfully. When I change the abunFilePath='normCoverageReduced.csv' to
abunFilePath='normCoverage.csv' , the error occurs that showed follow:
Error using fluxVariability (line 248)
The FVA could not be run because the model is infeasible or unbounded
Error in buildModelStorage (line 98)
[minFlux,maxFlux]=fluxVariability(model,0,'max',ex_rxns);
Error in mgPipe (line 101)
[activeExMets,couplingMatrix]=buildModelStorage(microbeNames, modPath, dietFilePath, adaptMedium,
includeHumanMets, numWorkers, pruneModels);
Error in initMgPipe (line 187)
[netSecretionFluxes, netUptakeFluxes, Y, modelStats, summary, statistics, modelsOK] = mgPipe(modPath,
abunFilePath, computeProfiles, resPath, dietFilePath, infoFilePath, hostPath, hostBiomassRxn, hostBiomassRxnFlux,
figForm, numWorkers, rDiet, pDiet, lowerBMBound, upperBMBound, includeHumanMets, adaptMedium, pruneModels);
The FVA could not be run because the model is infeasible or unbounded
Error in buildModelStorage (line 98)
[minFlux,maxFlux]=fluxVariability(model,0,'max',ex_rxns);
Error in mgPipe (line 101)
[activeExMets,couplingMatrix]=buildModelStorage(microbeNames, modPath, dietFilePath, adaptMedium,
includeHumanMets, numWorkers, pruneModels);
Error in initMgPipe (line 187)
[netSecretionFluxes, netUptakeFluxes, Y, modelStats, summary, statistics, modelsOK] = mgPipe(modPath,
abunFilePath, computeProfiles, resPath, dietFilePath, infoFilePath, hostPath, hostBiomassRxn, hostBiomassRxnFlux,
figForm, numWorkers, rDiet, pDiet, lowerBMBound, upperBMBound, includeHumanMets, adaptMedium, pruneModels);
the whole code I run as follow:

What should I do to resolve the problem?
Thank you
Sincerely!
Una
Almut Heinken
Mar 1, 2023, 5:34:12 AM3/1/23
to cobra-...@googlegroups.com
Dear Una,
could you try installing the IBM CPLEX solver and running fastFVA instead of fluxVariability?
Best regards,
Almut
--
---
You received this message because you are subscribed to the Google Groups "COBRA Toolbox" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cobra-toolbo...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/cobra-toolbox/a3f2d583-27e8-42f5-935f-2787a4ea1cecn%40googlegroups.com.
Reply all
Reply to author
Forward
0 new messages
