Comparing gene signatures
MSK INTERN
debr...@mskcc.org
Hi Alan,
Thanks for reaching out!
We currently do not allow for searching by gene signatures, but we are working on enabling that feature
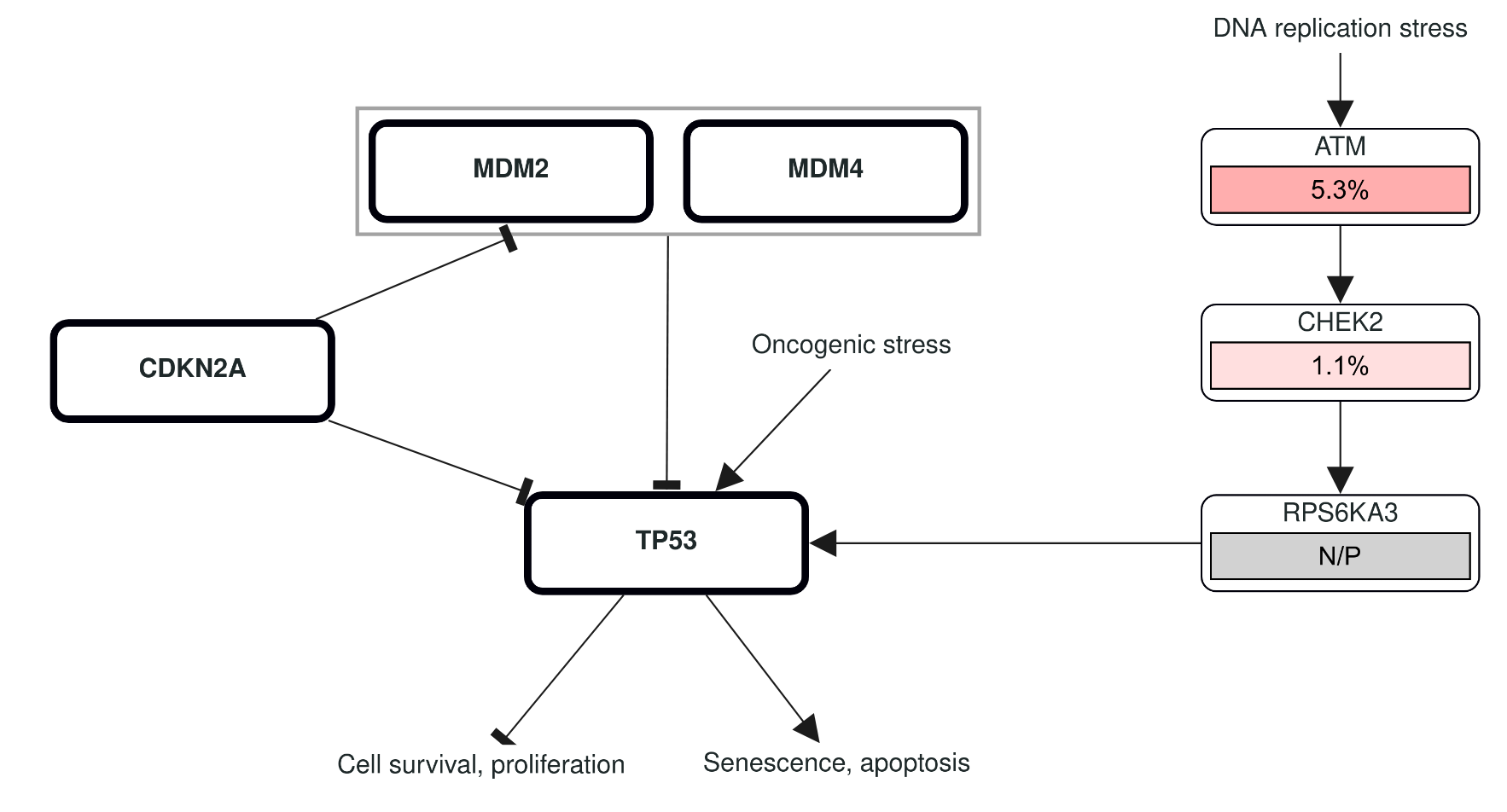
Note that on the results view it is already possible to add a “Mutation Spectrum” track to the Oncoprint so you can get an idea of the type of base changes in a sample. See attached screenshot
Hope that helps!
Best wishes,
Ino
--
You received this message because you are subscribed to the Google Groups "cBioPortal for Cancer Genomics Discussion Group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to
cbioportal+...@googlegroups.com.
To view this discussion on the web visit
https://groups.google.com/d/msgid/cbioportal/ef515aa9-7645-45f7-901e-cff581fa8012n%40googlegroups.com.
*** Only open attachments or links from trusted senders. Report phishing to inf...@mskcc.org ***
=====================================================================
Please note that this e-mail and any files transmitted from
Memorial Sloan Kettering Cancer Center may be privileged, confidential,
and protected from disclosure under applicable law. If the reader of
this message is not the intended recipient, or an employee or agent
responsible for delivering this message to the intended recipient,
you are hereby notified that any reading, dissemination, distribution,
copying, or other use of this communication or any of its attachments
is strictly prohibited. If you have received this communication in
error, please notify the sender immediately by replying to this message
and deleting this message, any attachments, and all copies and backups
from your computer.
debr...@mskcc.org
Hi Alan,
Apologies. I misread mutational signatures, rather than gene expression signatures
That said, we do not provide a way to query by that either in the public portal. If you are using a local version however it is possible to query and cluster by e.g. gene sets from MSigDB. CC’ing Sjoerd in case he wants to add anything
See also:
https://www.thehyve.nl/articles/gene-set-visualization-in-cbioportal-oncoprint
Thank you!
Ino
To view this discussion on the web visit
https://groups.google.com/d/msgid/cbioportal/2884CA43-5F20-49A7-A00B-274A9B4AB06A%40mskcc.org.
Sjoerd van Hagen