Issue searching gene expression
Alexandra Nicole Demetriou
Hello,
I’m trying to use cBioPortal to look at expression of a particular gene across brain tumor cases, and I watched the “cBioPortal: Comparing samples based on expression level of a gene” YouTube video for reference. When I search my gene of interest under Charts > Gene Specific, it says “unable to validate gene symbols.” Under “mutated genes,” I see that the gene is mutated in 4 cases. Does this mean that expression data is not available for that gene? Or is there another way I might assess expression levels across samples?
Thanks,
Alexandra
JJ Gao
--
You received this message because you are subscribed to the Google Groups "cBioPortal for Cancer Genomics Discussion Group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cbioportal+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/cbioportal/BYAPR07MB6072F67F97C95C3A9976DFA1C58F9%40BYAPR07MB6072.namprd07.prod.outlook.com.
Alexandra Nicole Demetriou
Hi JJ,
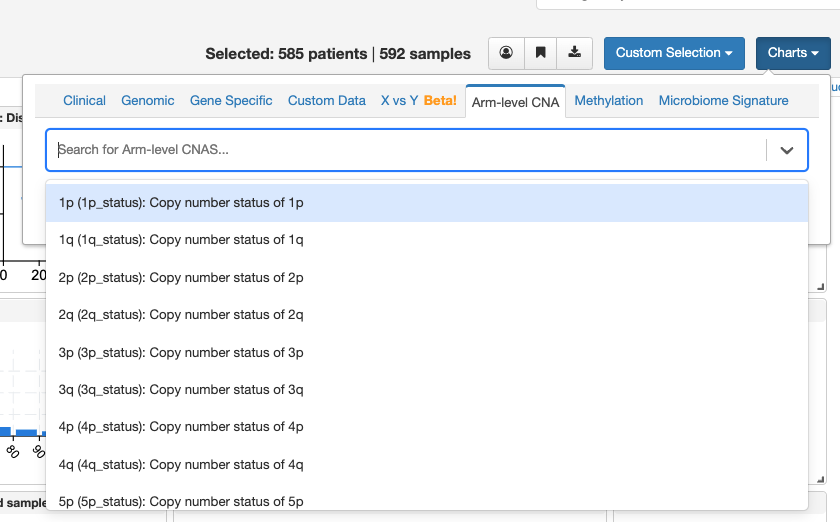
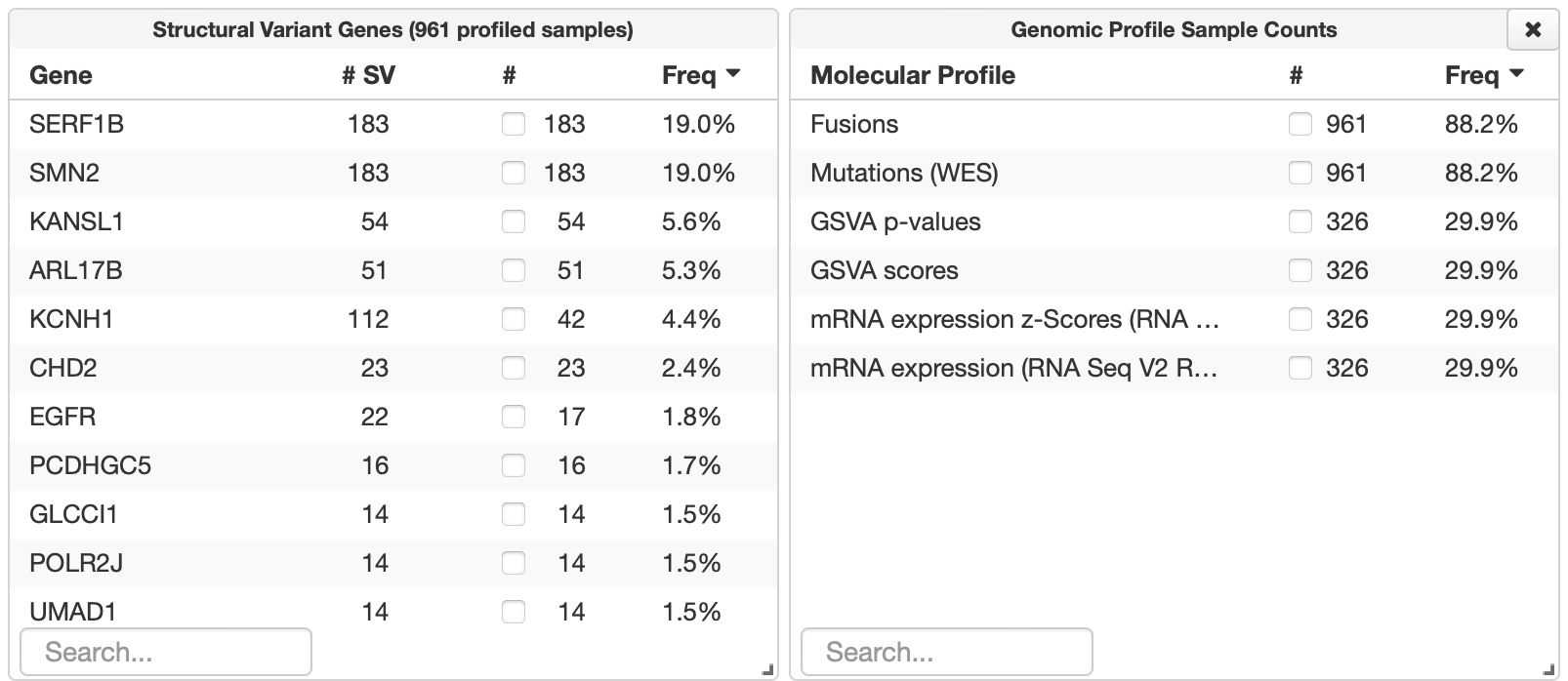
Thanks for getting back to me. I tried accessing cBioPortal through a different browser and it worked this time. I actually had another question – I’m trying to also look at gain and loss of entire chromosomes and I was having trouble doing so. I have a private dataset and I’m not sure if that makes a difference for what I can/can’t view in cBioPortal. My cBioPortal displays the following genomic data chart options (first screenshot), and when I display structural variant genes, the following comes up (second screenshot). I want to see cases that have gain of entire chromosome 7, for example, and I’m not sure if that’s possible here. Do you have any thoughts on how I might see that data in cBioPortal, or if it’s maybe not possible based on what I have here?
Thanks,
Alexandra

JJ Gao