Performance issiues using vanilla-biometrics
18 views
Skip to first unread message
teodors...@gmail.com
Feb 19, 2021, 6:37:19 AM2/19/21
to bob-devel
Hi again,
I am aware that the package is not fully developed; I only want to let you know that there are some performance issues.
Task:
Reproduce UTFVP baseline experiments;
Installation method:
built locally, using commands:
conda create -n bdt -c https://www.idiap.ch/software/bob/conda bob bob.devtools
[activate bdt]
git clone https://gitlab.idiap.ch/bob/bob.bio.vein
cd ./bob.bio.vein
bdt create -vv VEINS_DEV
[activate VEINS_DEV]
buildout
Version
Indicated version in the ``version.txt``: 1.1.7b0
Issiues
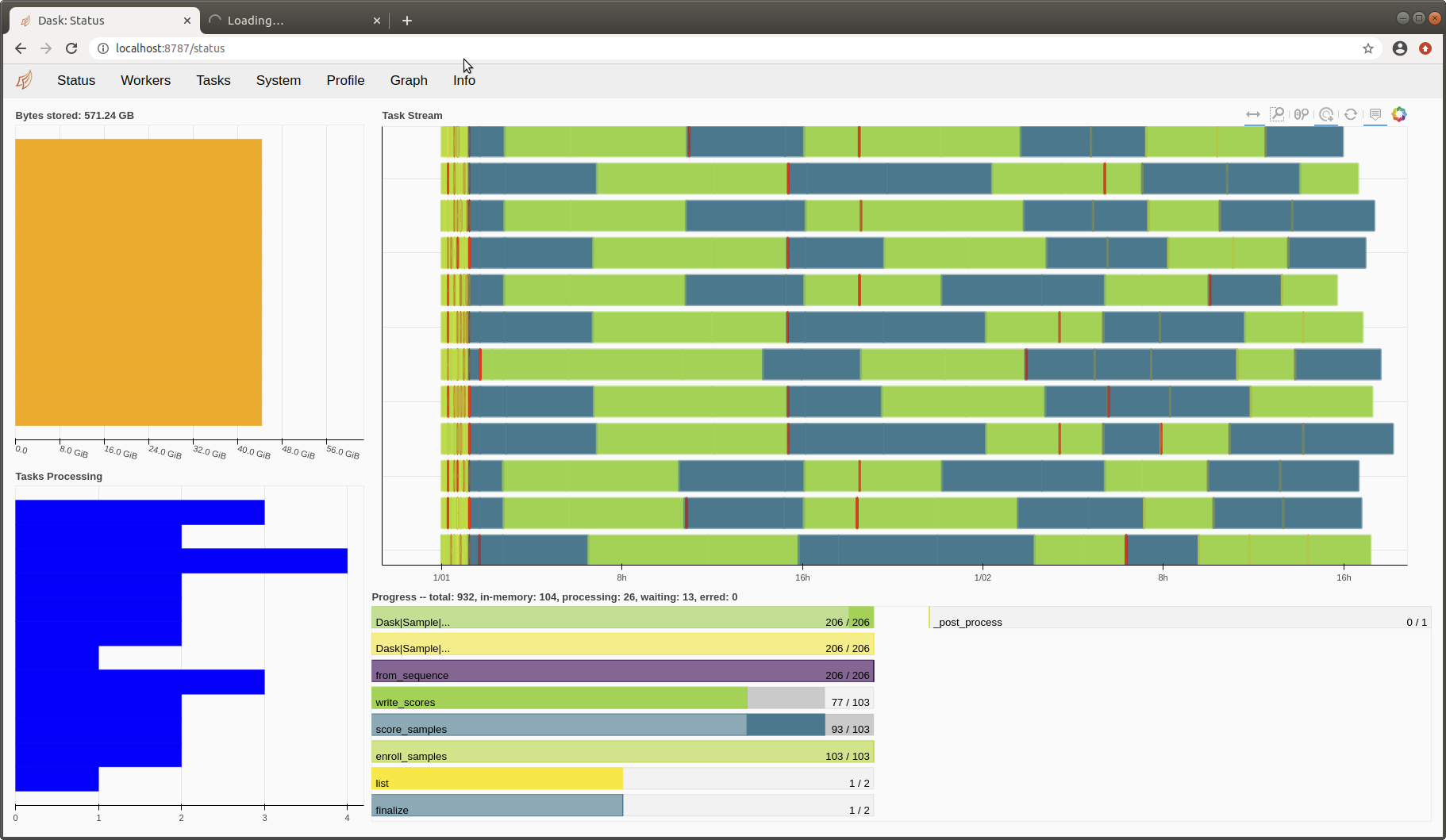
Experiments for the NOM dataset took some 15 hours for each -- dev and eval subset, using the command:
% ./bin/bob bio pipelines vanilla-biometrics nom mc utfvp -g eval -l local-parallel -o NOM_EVAL -vv
Experiments for the NOM dataset took some 15 hours for each -- dev and eval subset, using the command:
% ./bin/bob bio pipelines vanilla-biometrics nom mc utfvp -g eval -l local-parallel -o NOM_EVAL -vv
There were plenty of error messages during the execution (about delayed process execution for a specific core), but eventually, experiments finished producing the combined score file.
However, for ALL protocol:
% ./bin/bob bio pipelines vanilla-biometrics full utfvp mc -l local-parallel -o ALL -vv
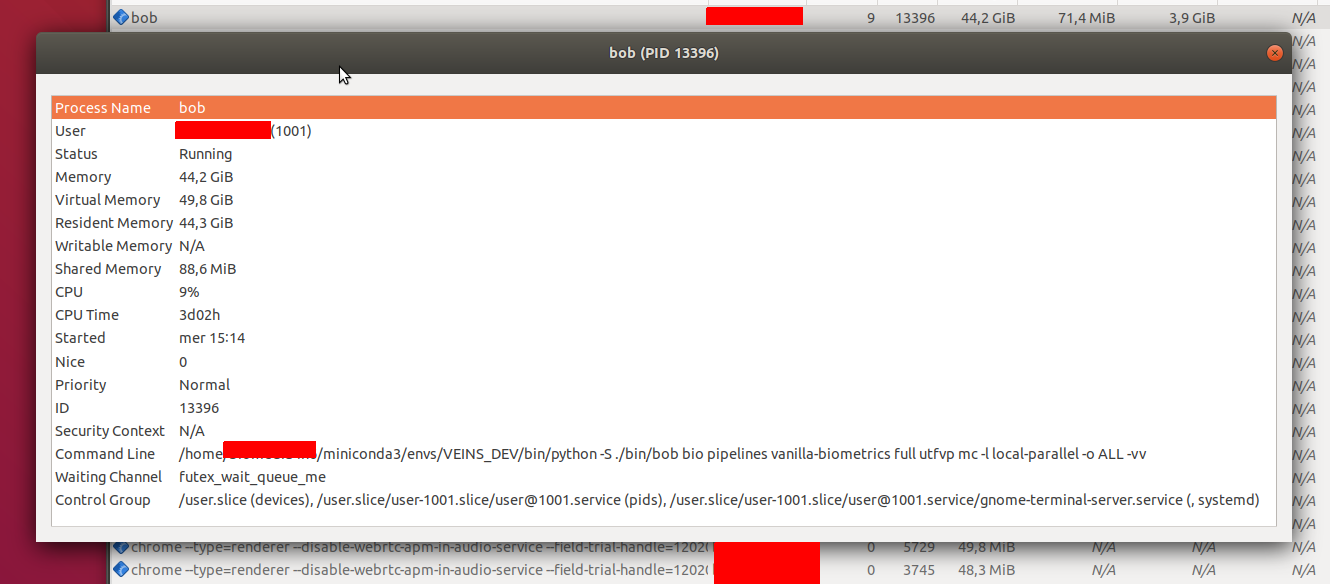
I had to stop execution after three days; eventually, the terminal was paralysed with messages like this:
distributed.worker - WARNING - Memory use is high but worker has no data to store to disk. Perhaps some other process is leaking memory? Process memory: 47.60 GB -- Worker memory limit: 67.36 GB
Hope this helps. Will proceed using the stable version. One of the main misunderstandings arises from the fact that if one google e.g. bob.bio.vein, more often than not the master branch version of the docs is the first result, instead of the stable version.
Hope this helps,
Teo
Some screenshots:
Tiago Freitas Pereira
Feb 19, 2021, 7:12:18 AM2/19/21
to bob-...@googlegroups.com
Hi Teodors,
Glad to hear that there's something testing `bob.bio.vein` :-)Second, to avoid the memory increase, add the option `--checkpoint` in your command.
Otherwise, all your experiment will be kept in memory until the score writing.
Let me know how it goes
Thanks
Tiago
--
-- You received this message because you are subscribed to the Google Groups bob-devel group. To post to this group, send email to bob-...@googlegroups.com. To unsubscribe from this group, send email to bob-devel+...@googlegroups.com. For more options, visit this group at https://groups.google.com/d/forum/bob-devel or directly the project website at http://idiap.github.com/bob/
---
You received this message because you are subscribed to the Google Groups "bob-devel" group.
To unsubscribe from this group and stop receiving emails from it, send an email to bob-devel+...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/bob-devel/34ee508d-a556-4ffa-9c1c-8d1e019b2cd2n%40googlegroups.com.
--
Tiago
Tiago Freitas Pereira
Feb 19, 2021, 7:18:48 AM2/19/21
to bob-...@googlegroups.com
Hi Teodors,
I'm requesting access to the UTFVP dataset, so I can test this with you.
Cheers
Tiago
Tiago
teodors...@gmail.com
Feb 19, 2021, 3:27:28 PM2/19/21
to bob-devel
Hi,
Today, with the stable version, nom protocol, both group dev and eval experiment execution time was about 2 hours; full took some four.
As I mentioned before, there were error messages constantly; but I didn't save them. If interest for you, can reproduce it; I don't have any clue about the fancy software version/implementation details.
Best,
Teo
Reply all
Reply to author
Forward
0 new messages
