structural equation value zero (p-value 1)
43 views
Skip to first unread message
TOMAS ECHIBURU
Mar 14, 2022, 3:39:45 AM3/14/22
to Biogeme
Dear Prof. Bierlaire,
No sienta la obligación de contestar este mail fuera de horario laboral.
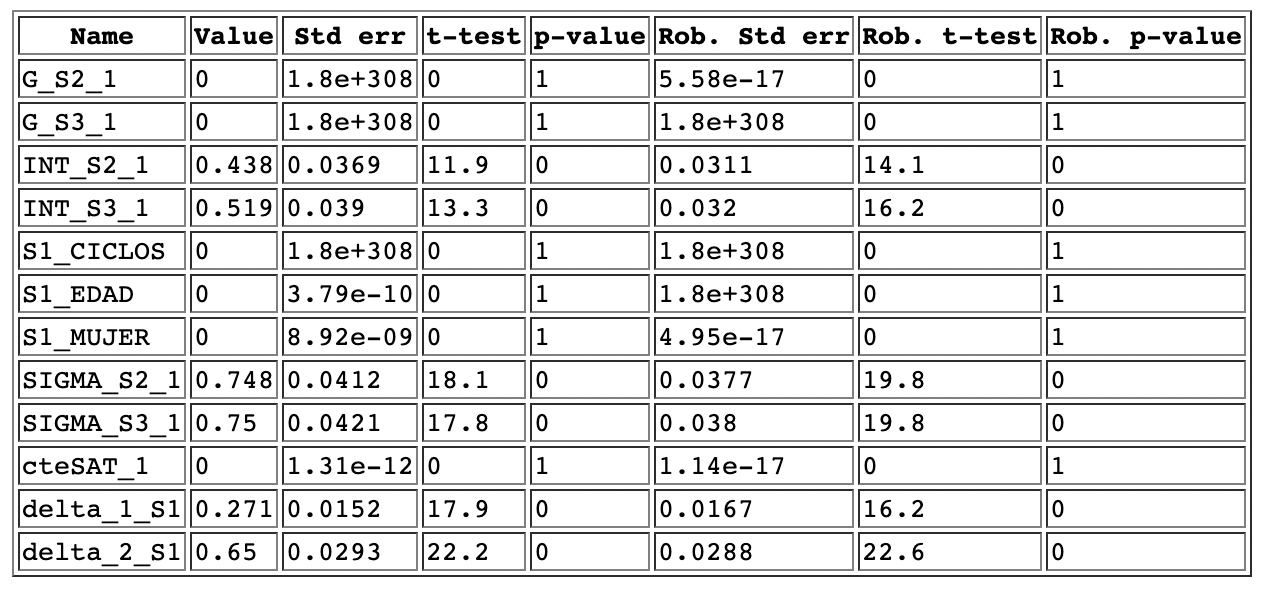
Im trying to estimate a latent variable model but some values are not being estiamted. Originally it had three latent variables, and I had this error. Then, for simplicity, I tried with only one LV but the issue persists: It seems that is not able to estimate values for the structural equation and, therefore, the parameter of the measurement equation that interacts with the utility function is also zero.
Any idea why this may be happening? Thank you so much for your time.

*** Here i'm attaching the script which is very simple.
import pandas as pd
import biogeme.database as db
import biogeme.biogeme as bio
from biogeme import models
import biogeme.messaging as msg
from biogeme.expressions import Beta, DefineVariable, log, Elem, bioNormalCdf
import biogeme.database as db
df = pd.read_csv('dfw.csv', sep=';')
database = db.Database('dfw', df)
globals().update(database.variables)
### VARIABLES
MUJER_1 = DefineVariable('MUJER_1', GEN_N_1 == 1, database)
MUJER_2 = DefineVariable('MUJER_2', GEN_N_2 == 1, database)
### INDICADORES Variables Latentes
### Satisfación 2018
S1_1_R = DefineVariable('S1_1_R', S3_1, database) # DISCRETA DE 1 A 5
S2_1_R = DefineVariable('S2_1_R', S4_1, database) # DISCRETA DE 1 A 5
S3_1_R = DefineVariable('S3_1_R', S5_1, database) # DISCRETA DE 1 A 5
########## PARAMETROS ECUACION ESTRUCTURAL ######################
cteSAT_1 = Beta('cteSAT_1', 0, None, None, 0)
S1_MUJER = Beta('S1_MUJER', 0, None, None, 0)
S1_EDAD = Beta('S1_EDAD', 0, None, None, 0)
S1_CICLOS = Beta('S1_CICLOS', 0, None, None, 0)
###################### ECUACION ESTRUCTURAL: VARIABLES LATENTES ######################
SAT_1 = (
cteSAT_1 +
S1_MUJER * MUJER_1 +
S1_CICLOS * CICLO_D_1 +
S1_EDAD * AGE_1
)
#### PARAMETROS ECUACION DE MEDICION ####
# ### SAT 2018
INT_S1_1 = Beta('INT_S1_1', 0, None, None, 1) ## FIJA EN 1
INT_S2_1 = Beta('INT_S2_1', 0, None, None, 0)
INT_S3_1 = Beta('INT_S3_1', 0, None, None, 0)
G_S1_1 = Beta('G_S1_1', 0, None, None, 1) ## FIJA EN 1
G_S2_1 = Beta('G_S2_1', 0, None, None, 0)
G_S3_1 = Beta('G_S3_1', 0, None, None, 0)
MOD_S1_1 = INT_S1_1 + G_S1_1 * SAT_1
MOD_S2_1 = INT_S2_1 + G_S2_1 * SAT_1
MOD_S3_1 = INT_S3_1 + G_S3_1 * SAT_1
SIGMA_S1_1 = Beta('SIGMA_S1_1', 1, None, None, 1) ## FIJA EN 1
SIGMA_S2_1 = Beta('SIGMA_S2_1', 1, None, None, 0)
SIGMA_S3_1 = Beta('SIGMA_S3_1', 1, None, None, 0)
# ### SAT 1
delta_1_S1 = Beta('delta_1_S1',0.1, 1.0e-5, None, 0)
delta_2_S1 = Beta('delta_2_S1',0.2, 1.0e-5, None, 0)
tau_1_S1 = -delta_1_S1 - delta_2_S1
tau_2_S1 = -delta_1_S1
tau_3_S1 = delta_1_S1
tau_4_S1 = delta_1_S1 + delta_2_S1
############## PROBABILIDADES ASOCIADAS A CADA ELECCION #########
#Se calcula la probabilidad que el modelo le asigna a la eleccion real
### Expedito 2018
S1_1_tau_1 = (tau_1_S1 - MOD_S1_1) / SIGMA_S1_1
S1_1_tau_2 = (tau_2_S1 - MOD_S1_1) / SIGMA_S1_1
S1_1_tau_3 = (tau_3_S1 - MOD_S1_1) / SIGMA_S1_1
S1_1_tau_4 = (tau_4_S1 - MOD_S1_1) / SIGMA_S1_1
IND_S1_1_M = {
1: bioNormalCdf(S1_1_tau_1),
2: bioNormalCdf(S1_1_tau_2) - bioNormalCdf(S1_1_tau_1),
3: bioNormalCdf(S1_1_tau_3) - bioNormalCdf(S1_1_tau_2),
4: bioNormalCdf(S1_1_tau_4) - bioNormalCdf(S1_1_tau_3),
5: 1 - bioNormalCdf(S1_1_tau_4),
}
P_S1_1 = Elem(IND_S1_1_M, S1_1_R)
# ### Agradable 2018
S2_1_tau_1 = (tau_1_S1 - MOD_S2_1) / SIGMA_S2_1
S2_1_tau_2 = (tau_2_S1 - MOD_S2_1) / SIGMA_S2_1
S2_1_tau_3 = (tau_3_S1 - MOD_S2_1) / SIGMA_S2_1
S2_1_tau_4 = (tau_4_S1 - MOD_S2_1) / SIGMA_S2_1
IND_S2_1_M = {
1: bioNormalCdf(S2_1_tau_1),
2: bioNormalCdf(S2_1_tau_2) - bioNormalCdf(S2_1_tau_1),
3: bioNormalCdf(S2_1_tau_3) - bioNormalCdf(S2_1_tau_2),
4: bioNormalCdf(S2_1_tau_4) - bioNormalCdf(S2_1_tau_3),
5: 1 - bioNormalCdf(S2_1_tau_4),
}
P_S2_1 = Elem(IND_S2_1_M, S2_1_R)
## Satisfecho 2018
S3_1_tau_1 = (tau_1_S1 - MOD_S3_1) / SIGMA_S3_1
S3_1_tau_2 = (tau_2_S1 - MOD_S3_1) / SIGMA_S3_1
S3_1_tau_3 = (tau_3_S1 - MOD_S3_1) / SIGMA_S3_1
S3_1_tau_4 = (tau_4_S1 - MOD_S3_1) / SIGMA_S3_1
IND_S3_1_M = {
1: bioNormalCdf(S3_1_tau_1),
2: bioNormalCdf(S3_1_tau_2) - bioNormalCdf(S3_1_tau_1),
3: bioNormalCdf(S3_1_tau_3) - bioNormalCdf(S3_1_tau_2),
4: bioNormalCdf(S3_1_tau_4) - bioNormalCdf(S3_1_tau_3),
5: 1 - bioNormalCdf(S3_1_tau_4),
}
P_S3_1 = Elem(IND_S3_1_M, S3_1_R)
#########################################
loglike = (
log(P_S1_1) +
log(P_S2_1) +
log(P_S3_1)
)
# Define level of verbosity
logger = msg.bioMessage()
# logger.setSilent()
# logger.setWarning()
logger.setGeneral()
# logger.setDetailed()
# Create the Biogeme object
biogeme = bio.BIOGEME(database, loglike, numberOfDraws=20000)
biogeme.modelName = 'LV_S1'
# Estimate the parameters
results = biogeme.estimate()
print(f'Estimated betas: {len(results.data.betaValues)}')
print(f'final log likelihood: {results.data.logLike:.3f}')
print(f'Output file: {results.data.htmlFileName}')
results.writeLaTeX()
print(f'LaTeX file: {results.data.latexFileName}')
import biogeme.database as db
import biogeme.biogeme as bio
from biogeme import models
import biogeme.messaging as msg
from biogeme.expressions import Beta, DefineVariable, log, Elem, bioNormalCdf
import biogeme.database as db
df = pd.read_csv('dfw.csv', sep=';')
database = db.Database('dfw', df)
globals().update(database.variables)
### VARIABLES
MUJER_1 = DefineVariable('MUJER_1', GEN_N_1 == 1, database)
MUJER_2 = DefineVariable('MUJER_2', GEN_N_2 == 1, database)
### INDICADORES Variables Latentes
### Satisfación 2018
S1_1_R = DefineVariable('S1_1_R', S3_1, database) # DISCRETA DE 1 A 5
S2_1_R = DefineVariable('S2_1_R', S4_1, database) # DISCRETA DE 1 A 5
S3_1_R = DefineVariable('S3_1_R', S5_1, database) # DISCRETA DE 1 A 5
########## PARAMETROS ECUACION ESTRUCTURAL ######################
cteSAT_1 = Beta('cteSAT_1', 0, None, None, 0)
S1_MUJER = Beta('S1_MUJER', 0, None, None, 0)
S1_EDAD = Beta('S1_EDAD', 0, None, None, 0)
S1_CICLOS = Beta('S1_CICLOS', 0, None, None, 0)
###################### ECUACION ESTRUCTURAL: VARIABLES LATENTES ######################
SAT_1 = (
cteSAT_1 +
S1_MUJER * MUJER_1 +
S1_CICLOS * CICLO_D_1 +
S1_EDAD * AGE_1
)
#### PARAMETROS ECUACION DE MEDICION ####
# ### SAT 2018
INT_S1_1 = Beta('INT_S1_1', 0, None, None, 1) ## FIJA EN 1
INT_S2_1 = Beta('INT_S2_1', 0, None, None, 0)
INT_S3_1 = Beta('INT_S3_1', 0, None, None, 0)
G_S1_1 = Beta('G_S1_1', 0, None, None, 1) ## FIJA EN 1
G_S2_1 = Beta('G_S2_1', 0, None, None, 0)
G_S3_1 = Beta('G_S3_1', 0, None, None, 0)
MOD_S1_1 = INT_S1_1 + G_S1_1 * SAT_1
MOD_S2_1 = INT_S2_1 + G_S2_1 * SAT_1
MOD_S3_1 = INT_S3_1 + G_S3_1 * SAT_1
SIGMA_S1_1 = Beta('SIGMA_S1_1', 1, None, None, 1) ## FIJA EN 1
SIGMA_S2_1 = Beta('SIGMA_S2_1', 1, None, None, 0)
SIGMA_S3_1 = Beta('SIGMA_S3_1', 1, None, None, 0)
# ### SAT 1
delta_1_S1 = Beta('delta_1_S1',0.1, 1.0e-5, None, 0)
delta_2_S1 = Beta('delta_2_S1',0.2, 1.0e-5, None, 0)
tau_1_S1 = -delta_1_S1 - delta_2_S1
tau_2_S1 = -delta_1_S1
tau_3_S1 = delta_1_S1
tau_4_S1 = delta_1_S1 + delta_2_S1
############## PROBABILIDADES ASOCIADAS A CADA ELECCION #########
#Se calcula la probabilidad que el modelo le asigna a la eleccion real
### Expedito 2018
S1_1_tau_1 = (tau_1_S1 - MOD_S1_1) / SIGMA_S1_1
S1_1_tau_2 = (tau_2_S1 - MOD_S1_1) / SIGMA_S1_1
S1_1_tau_3 = (tau_3_S1 - MOD_S1_1) / SIGMA_S1_1
S1_1_tau_4 = (tau_4_S1 - MOD_S1_1) / SIGMA_S1_1
IND_S1_1_M = {
1: bioNormalCdf(S1_1_tau_1),
2: bioNormalCdf(S1_1_tau_2) - bioNormalCdf(S1_1_tau_1),
3: bioNormalCdf(S1_1_tau_3) - bioNormalCdf(S1_1_tau_2),
4: bioNormalCdf(S1_1_tau_4) - bioNormalCdf(S1_1_tau_3),
5: 1 - bioNormalCdf(S1_1_tau_4),
}
P_S1_1 = Elem(IND_S1_1_M, S1_1_R)
# ### Agradable 2018
S2_1_tau_1 = (tau_1_S1 - MOD_S2_1) / SIGMA_S2_1
S2_1_tau_2 = (tau_2_S1 - MOD_S2_1) / SIGMA_S2_1
S2_1_tau_3 = (tau_3_S1 - MOD_S2_1) / SIGMA_S2_1
S2_1_tau_4 = (tau_4_S1 - MOD_S2_1) / SIGMA_S2_1
IND_S2_1_M = {
1: bioNormalCdf(S2_1_tau_1),
2: bioNormalCdf(S2_1_tau_2) - bioNormalCdf(S2_1_tau_1),
3: bioNormalCdf(S2_1_tau_3) - bioNormalCdf(S2_1_tau_2),
4: bioNormalCdf(S2_1_tau_4) - bioNormalCdf(S2_1_tau_3),
5: 1 - bioNormalCdf(S2_1_tau_4),
}
P_S2_1 = Elem(IND_S2_1_M, S2_1_R)
## Satisfecho 2018
S3_1_tau_1 = (tau_1_S1 - MOD_S3_1) / SIGMA_S3_1
S3_1_tau_2 = (tau_2_S1 - MOD_S3_1) / SIGMA_S3_1
S3_1_tau_3 = (tau_3_S1 - MOD_S3_1) / SIGMA_S3_1
S3_1_tau_4 = (tau_4_S1 - MOD_S3_1) / SIGMA_S3_1
IND_S3_1_M = {
1: bioNormalCdf(S3_1_tau_1),
2: bioNormalCdf(S3_1_tau_2) - bioNormalCdf(S3_1_tau_1),
3: bioNormalCdf(S3_1_tau_3) - bioNormalCdf(S3_1_tau_2),
4: bioNormalCdf(S3_1_tau_4) - bioNormalCdf(S3_1_tau_3),
5: 1 - bioNormalCdf(S3_1_tau_4),
}
P_S3_1 = Elem(IND_S3_1_M, S3_1_R)
#########################################
loglike = (
log(P_S1_1) +
log(P_S2_1) +
log(P_S3_1)
)
# Define level of verbosity
logger = msg.bioMessage()
# logger.setSilent()
# logger.setWarning()
logger.setGeneral()
# logger.setDetailed()
# Create the Biogeme object
biogeme = bio.BIOGEME(database, loglike, numberOfDraws=20000)
biogeme.modelName = 'LV_S1'
# Estimate the parameters
results = biogeme.estimate()
print(f'Estimated betas: {len(results.data.betaValues)}')
print(f'final log likelihood: {results.data.logLike:.3f}')
print(f'Output file: {results.data.htmlFileName}')
results.writeLaTeX()
print(f'LaTeX file: {results.data.latexFileName}')
No sienta la obligación de contestar este mail fuera de horario laboral.
Dirección de Personas UC
Bierlaire Michel
Mar 14, 2022, 3:45:23 AM3/14/22
to tomas.e...@uc.cl, Bierlaire Michel, Biogeme
Apparently, you did not include the measurement equations in the loglikelihood function.
On 13 Mar 2022, at 16:42, 'TOMAS ECHIBURU' via Biogeme <bio...@googlegroups.com> wrote:
Dear Prof. Bierlaire,
Im trying to estimate a latent variable model but some values are not being estiamted. Originally it had three latent variables, and I had this error. Then, for simplicity, I tried with only one LV but the issue persists: It seems that is not able to estimate values for the structural equation and, therefore, the parameter of the measurement equation that interacts with the utility function is also zero.Any idea why this may be happening? Thank you so much for your time.
--
You received this message because you are subscribed to the Google Groups "Biogeme" group.
To unsubscribe from this group and stop receiving emails from it, send an email to biogeme+u...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/biogeme/0637f0af-fc4e-4d77-8f3a-9557b57ec1c7n%40googlegroups.com.
Reply all
Reply to author
Forward
0 new messages
