ImportError: DLL load failed while importing cythonbiogeme: The specified module could not be found.
2,483 views
Skip to first unread message
oz zalel
Aug 28, 2022, 1:06:59 PM8/28/22
to Biogeme
Hi
michel,
After installing Biogeme 3.2.10 using pip 21.1.2 I got the following error
ImportError: DLL load failed while importing cythonbiogeme: The specified module could not be found.
this is the code I was trying to run:
import pandas as pd
from matplotlib import pyplot as plt
from matplotlib import pyplot as plt
import biogeme.database as db
import biogeme.biogeme as bio
from biogeme.expressions import Beta, DefineVariable
from biogeme.models import loglogit.
import biogeme.biogeme as bio
from biogeme.expressions import Beta, DefineVariable
from biogeme.models import loglogit.
I'll be happy for your help about it.
thank you very much
Bierlaire Michel
Aug 28, 2022, 1:10:35 PM8/28/22
to zale...@gmail.com, Bierlaire Michel, Biogeme
Which version of python are you using?
Install cython, pandas, scipy and tqdm.
Then install biogeme.
Let me know if it works.
--
You received this message because you are subscribed to the Google Groups "Biogeme" group.
To unsubscribe from this group and stop receiving emails from it, send an email to biogeme+u...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/biogeme/b3b7c26b-82cf-4d0e-b034-2055ffdb5481n%40googlegroups.com.
André Duarte
Sep 6, 2022, 11:27:30 AM9/6/22
to Biogeme
Hi.
I had a similar problem. Import DLL error for
cythonbiogem, then biogeme.database and finaly this error.
------
Exception has occurred: ImportError
Unable to import required dependencies:
numpy:
IMPORTANT: PLEASE READ THIS FOR ADVICE ON HOW TO SOLVE THIS ISSUE!
Importing the numpy C-extensions failed. This error can happen for
many reasons, often due to issues with your setup or how NumPy was
installed.
We have compiled some common reasons and troubleshooting tips at:
https://numpy.org/devdocs/user/troubleshooting-importerror.html
Please note and check the following:
* The Python version is: Python3.9 from "C:\ProgramData\Anaconda3\python.exe"
* The NumPy version is: "1.21.5"
and make sure that they are the versions you expect.
Please carefully study the documentation linked above for further help.
Original error was: DLL load failed while importing _multiarray_umath: The specified module could not be found.
File :XXX\\DiscreteModelingCBB\v422_binary_SM_specificHeadwaySocioEC.py", line 9, in <module>
import pandas as pd
------
In a fresh install of a laptop, with Windows 10.
The only solution we found at the moment was to have the computer with linux on the background and having Windows 10 running on top of it.
I´m not expert on the matter, but I can put in contact with who helped to solve the issue.
André Duarte
Bierlaire Michel
Sep 6, 2022, 11:42:05 AM9/6/22
to andrecantoe...@gmail.com, Bierlaire Michel, Biogeme
It seems to be due to conflicting versions of numpy.
The following versions of numpy have been used to create the executables:
Python 3.10 —> numpy 1.23.2
Python 3.9 —> numpy 1.23.2
Python 3.8 —> numpy 1.23.2
Python 3.7 —> numpy 1.21.6
To view this discussion on the web visit https://groups.google.com/d/msgid/biogeme/e9b8386c-7e26-4f9f-a676-a5f3ffba58b3n%40googlegroups.com.
andrecantoe...@gmail.com
Sep 6, 2022, 11:57:21 AM9/6/22
to Bierlaire Michel, Biogeme
Maybe.
But it´s strange. Fresh install of windows 10. I Might have installed python or pandas via terminal and then anaconda.
The only solution we found, was to install WSL – Ubuntu to run Windows on top of linux
Bierlaire Michel
Sep 6, 2022, 11:59:01 AM9/6/22
to andrecantoe...@gmail.com, Bierlaire Michel, Biogeme
Yes, this is indeed strange.
You may also try to install from sources.
- Install MSYS2.
- Add c:\msys64\mingw64\bin in the Windows PATH.
- Install using the following command:
pip install --global-option build_ext --global-option --compiler=mingw32.
Gerben
Sep 11, 2022, 2:26:02 AM9/11/22
to Biogeme
Hi Michel and others,
We were having similar problems with Python 3.10 and Biogeme 3.2.10. For what it's worth, rolling back to Python 3.9 and Biogeme 3.2.8 helps as a temporary solution.
Best, Gerben
Op dinsdag 6 september 2022 om 18:59:01 UTC+3 schreef michel.b...@epfl.ch:
oz zalel
Oct 26, 2022, 8:20:45 AM10/26/22
to Biogeme
Hi Michel,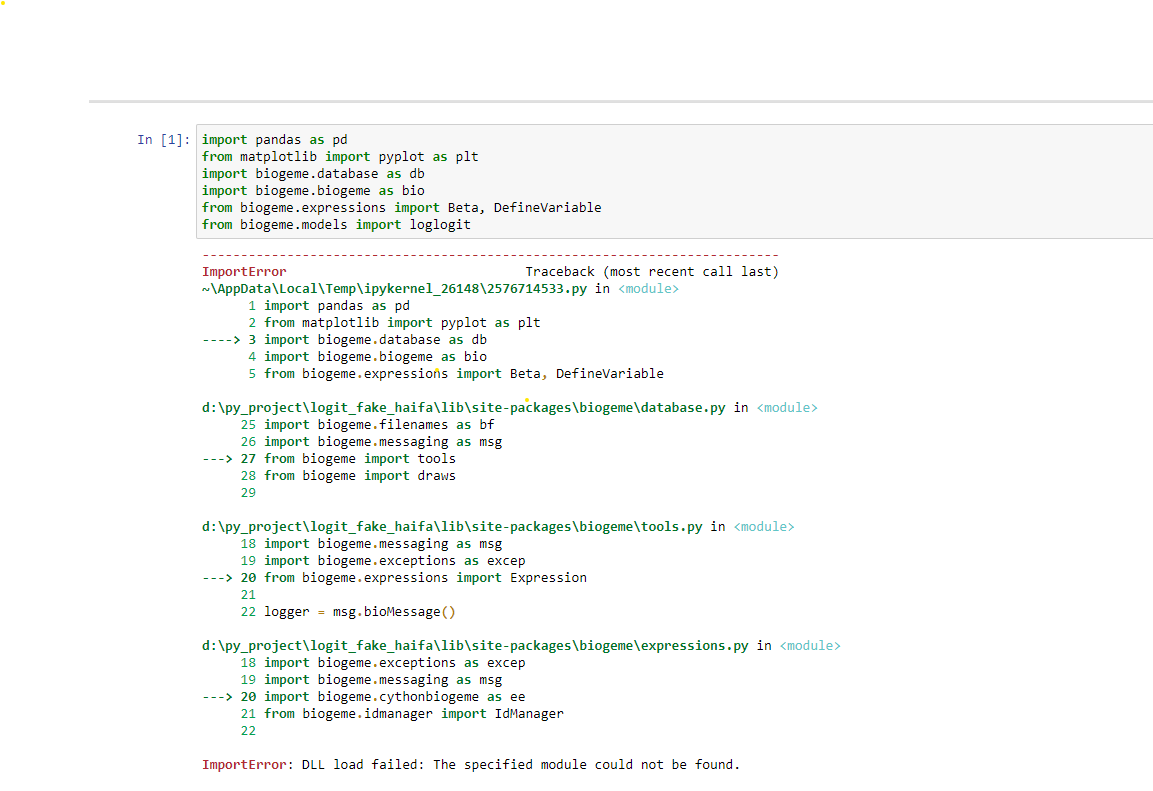
Thank you for the answer.
I've tried to create new
environment, installing to package in the order
I'm using python 3.7.6
cython 0.29.32
padnas 1.3.5
scipy 1.7.3
scipy 1.7.3
tqdm 4.64.1 and
biogeme 3.2.10
I'm still getting the following answer. I'll be happy for any other suggestion or advice regarding it

Bierlaire Michel
Oct 26, 2022, 8:22:42 AM10/26/22
to zale...@gmail.com, Bierlaire Michel, Biogeme
Very strange.
Why don’t you use a more recent version of python?
On 26 Oct 2022, at 14:16, oz zalel <zale...@gmail.com> wrote:
Hi Michel,Thank you for the answer.I've tried to create new environment, installing to package in the orderI'm using python 3.7.6cython 0.29.32padnas 1.3.5
scipy 1.7.3tqdm 4.64.1 andbiogeme 3.2.10I'm still getting the following answer. I'll be happy for any other suggestion or advice regarding it
To view this discussion on the web visit https://groups.google.com/d/msgid/biogeme/a7c2fa22-24ab-49db-8bba-65329812432fn%40googlegroups.com.
<biogeme_dll_error.png>
Cecilia Hult
Oct 27, 2022, 2:33:08 AM10/27/22
to Biogeme
Hi everyone, I also had this problem earlier today (first time installing Biogeme) with the following environment:
python 3.10.3
biogeme 3.2.10
python 3.10.3
biogeme 3.2.10
numpy 1.23.2 (downgraded from 1.23.4 according to Michels instructions earlier)
cython 0.29.32
scipy 1.9.3
tqdm 4.64.1
For me, the issue was a missing DLL (libwinpthread-1). I happened to have this DLL in my Julia lib and made a copy in the same folder as python.exe. After doing this, the "import biogeme..." worked fine. Have not had time yet to see if there are any subsequent issues.
oz zalel
Oct 27, 2022, 3:24:12 AM10/27/22
to Biogeme
Hi Michel,
installing version 3.2.8 is working so I guess this is the solution for me.
Thank you
Tom Eagle
Nov 1, 2022, 2:36:26 AM11/1/22
to Biogeme
I copied the file
libwinpthread-1, like
cecili...@ivl.se above suggested, from my folder 'C:\Program Files\Microsoft Visual Studio\2022\Community\Common7\IDE\CommonExtensions\Microsoft\TeamFoundation\Team Explorer\Git\mingw64\bin\libwinpthread-1.dll'. I copied it into my python directory 'C:\Users\'name'\AppData\Local\Programs\Python\Python39\Scripts'. After doing that biogeme 3.2.10 worked fine. As I was trying to run the 04LatentChoiceSeq.py program I found I also had to install the Jinja2 module using 'pip install Jinja2'
Facundo STORANI
Nov 25, 2022, 4:57:07 AM11/25/22
to Biogeme
I had the same issue while trying to run Biogeme on Windows.
"
import biogeme.cythonbiogeme as ee ImportError: DLL load failed while
importing cythonbiogeme: The specified module could not be found".
The issue is that in Python 3.8 or newer on Windows https://docs.python.org/3/whatsnew/3.8.html#ctypes
, files DLLs are loaded from trusted locations.
It is necessary to add
the path of the dlls to the directory:
- Search the DLLs folder of anaconda3. It may be similar to: C:\Users\[USER_NAME]\anaconda3\DLLs or C:\ProgramData\Anaconda3\DLLs
- Click the Start button, type “environment properties” into the search bar and hit Enter.
- In the System Properties window, click “Environment Variables.”
- Select "Path" on the users' list, and modify.
- Add the path of the dlls folder to the list.
Then I executed the cmd terminal and ran a code with no issues.
Tom Eagle
Dec 20, 2022, 4:50:11 PM12/20/22
to Biogeme
I screwed up the directory to put libwinpthread-1.dll in. it should go in your main Python directory. in my case it was
'C:\Users\'name'\AppData\Local\Programs\Python\Python39' not
'C:\Users\'name'\AppData\Local\Programs\Python\Python39\Scripts. Biogeme worked fine without adding any paths.
Salma Medhat
Apr 4, 2023, 11:28:28 AM4/4/23
to Biogeme
Hi everyone,
I am facing the same error and tried the following fixes:
1. Adding the path of the DLL folder to the Path variable (attached below is a screenshot).
2. I created a virtual environment and installed the requirements with the following versions:
biogeme 3.2.10
cython 0.29.34
numpy 1.23.5
scipy 1.10.1
tqdm 4.65.0
python 3.10.10
3. I intstalled Microsoft Visual C++
4. I tried coping the libwinpthread-1.dll file from 'C:\Program Files\Microsoft Visual Studio\2022\Professional\Common7\IDE\CommonExtensions\Microsoft\TeamFoundation\Team Explorer\Git\mingw64\bin' to 'C:\Users\User\anaconda3\envs\myenv_biogeme\Scripts'
However, the error persisted, could you kindly suggest a solution?
Best regards,
Salma Bauomy
Mat Guy
Apr 19, 2023, 8:20:46 AM4/19/23
to Biogeme
Hello Michel,
I have the same problem as the others.
Nonetheless, when I do this, the error changes into: "No module named 'biogeme.database''" but I can not install this module apart since it gives the following error:
"ERROR: Could not find a version that satisfies the requirement biogeme.database (from versions: none)
ERROR: No matching distribution found for biogeme.database"...
Could someone help me please?
Best regards,
M G
Bierlaire Michel
Apr 19, 2023, 8:23:05 AM4/19/23
to math...@orange.fr, Bierlaire Michel, Biogeme
Windows is a real mess. Everything works fine on Linux and MacOSX…
On the Windows computers where I tested the software before releasing it, it was working fine.
I am currently working on the next version of Biogeme. Hopefully, the issue will be fixed, although there is no guarantee.
> To view this discussion on the web visit https://groups.google.com/d/msgid/biogeme/513b79a7-872d-407d-a2da-f9c2e5a23de1n%40googlegroups.com.
Michel Bierlaire
Transport and Mobility Laboratory
School of Architecture, Civil and Environmental Engineering
EPFL - Ecole Polytechnique Fédérale de Lausanne
http://transp-or.epfl.ch
http://people.epfl.ch/michel.bierlaire
On the Windows computers where I tested the software before releasing it, it was working fine.
I am currently working on the next version of Biogeme. Hopefully, the issue will be fixed, although there is no guarantee.
Michel Bierlaire
Transport and Mobility Laboratory
School of Architecture, Civil and Environmental Engineering
EPFL - Ecole Polytechnique Fédérale de Lausanne
http://transp-or.epfl.ch
http://people.epfl.ch/michel.bierlaire
Mat Guy
Apr 19, 2023, 8:42:53 AM4/19/23
to Biogeme
Ok, but what is the best solution now if we have Python version 3.10.2 and Biogeme version 3.2.10 on Windows?
Best regards,
M G
Mat Guy
Apr 19, 2023, 10:57:41 AM4/19/23
to Biogeme
Hello,
I tried to do the same by downloading the DLL from the Internet (libwinpthread-1.dll free download | DLL‑files.com (dll-files.com)) since it was not present in the DLLs folder used by Python.
But now, I have the following problem: "ImportError: DLL load failed while importing cythonbiogeme: %1 n’est pas une application Win32 valide." (the end is "%1 is not a valid Win32 application"...).
Best regards,
M G
Bierlaire Michel
Apr 21, 2023, 2:14:03 AM4/21/23
to math...@orange.fr, Bierlaire Michel, Biogeme
Can you please try to install Biogeme 3.2.11?
pip install biogeme
Hopefully, the problem has been fixed.
> To view this discussion on the web visit https://groups.google.com/d/msgid/biogeme/63e1b8b8-8646-4d9d-bd21-e65980d6af4dn%40googlegroups.com.
pip install biogeme
Hopefully, the problem has been fixed.
Mat Guy
Apr 21, 2023, 9:43:06 AM4/21/23
to Biogeme
Yes, it works!
PS: Using Python v3.9.7 with Biogeme 3.2.10 also works, that's what I used yesterday.
Thank you!
Best regards,
M G
PS: Using Python v3.9.7 with Biogeme 3.2.10 also works, that's what I used yesterday.
Reply all
Reply to author
Forward
0 new messages
