some questions about unrooted tree
141 views
Skip to first unread message
苏秦之
Aug 5, 2019, 1:22:45 AM8/5/19
to ggtree
Hi, I have some questions about unrooted tree. My code is as follows.
treeg <- read.tree("genome_fasttree.txt")
groupg <- read.table("genome_tree_annotation.txt",header=T,sep="\t")
groupginfo_g <-split(groupg$strain,groupg$species)
treegg<-ggtree(treeg,layout="daylight",size=3,branch.length='none')
treegg_group<-groupOTU(treegg,groupginfo_g,'host')+aes(color=host)+
scale_color_manual(values=c("#1a1a1a","#f46d43","#3288bd","#1a9850","#c51b7d"))+
geom_tiplab(size=8)+
theme(legend.position="right")
pg<-treegg_group+new_scale_fill()
pg1<-pg+new_scale_color()
cluster<-groupg[,c(2,3)]
pg2<-pg1 %<+% cluster +
geom_tippoint(aes(color=cluster,shape=cluster),alpha=.7,shape=16)+
scale_color_manual(breaks=c("Gillia_Acer_1","Gillia_Acer_2","Gillia_Acer_3","Gillia_Acer_4","Gillia_Acer_5","none"),values=c("#66c2a5","#8da0cb","#e78ac3","#a6d854","#ffd92f","#737373"))+
scale_shape_manual(breaks=c("Gillia_Acer_1","Gillia_Acer_2","Gillia_Acer_3","Gillia_Acer_4","Gillia_Acer_5","NA"),values=c(21,22,23,24,25,21))
pg2
ggsave("genome_tree2.pdf",treegg_group,width=48,height=40)
I have two problems.
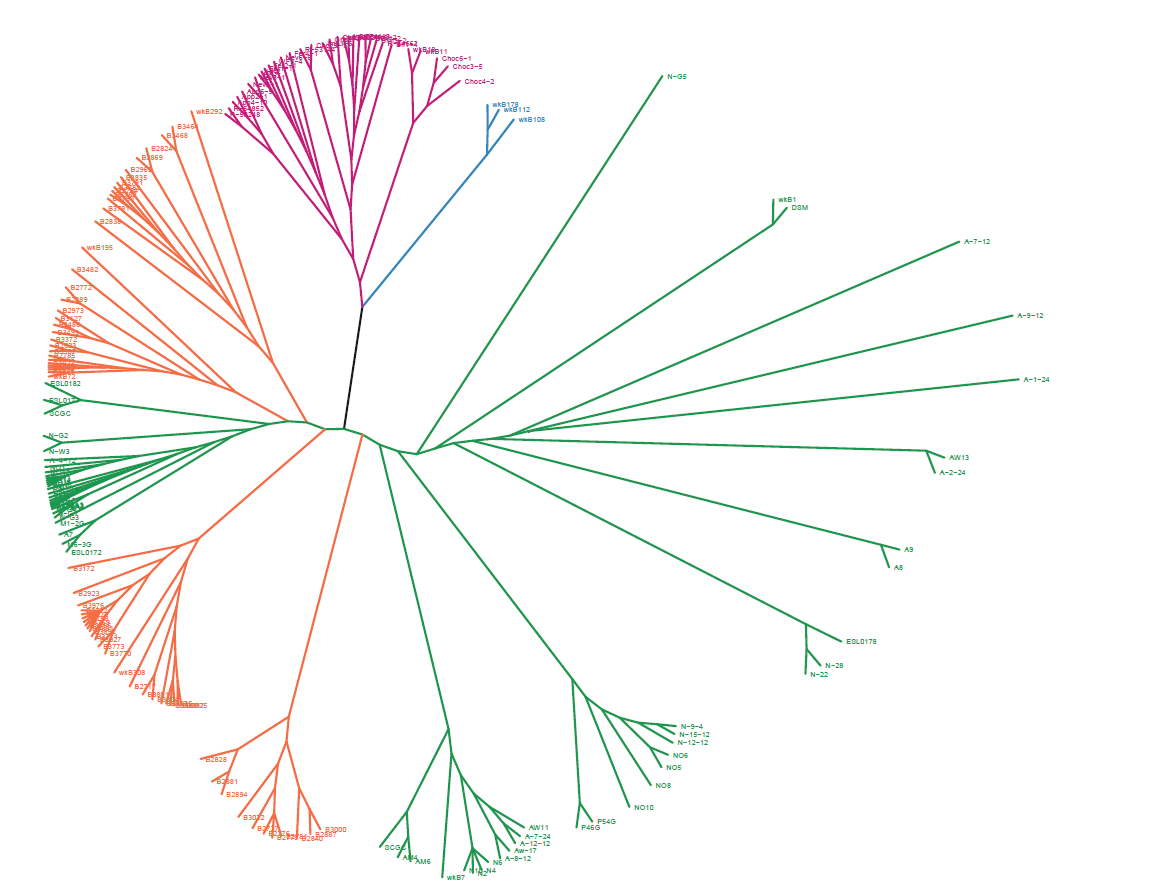
First, my tip label embedded in the branches as the picture.
Second, the error information as follows:
> pg2 错误: Insufficient values in manual scale. 11 needed but only 6 provided.
I don't know how to solve the problems. Thank you for your help.
Yu, Guangchuang
Aug 5, 2019, 3:18:17 AM8/5/19
to 苏秦之, ggtree
reproducible example pls.
--
1. G Yu*, TTY Lam, H Zhu, Y Guan*. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Molecular Biology and Evolution, 2018, 35(2):3041-3043. doi: 10.1093/molbev/msy194
2. G Yu, DK Smith, H Zhu, Y Guan, TTY Lam*. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data, Methods in Ecology and Evolution, 2017, 8(1):28-36. doi:10.1111/2041-210X.12628
3. Book: https://yulab-smu.github.io/treedata-book/
---
You received this message because you are subscribed to the Google Groups "ggtree" group.
To unsubscribe from this group and stop receiving emails from it, send an email to bioc-ggtree...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/bioc-ggtree/e902cf8a-c017-4e4c-afb1-bd5c2a91d6a1%40googlegroups.com.
--
--~--~---------~--~----~------------~-------~--~----~
Guangchuang Yu PhD
Professor
School of Basic Medical Sciences
School of Basic Medical Sciences
Southern Medical University
Guangzhou, China
-~----------~----~----~----~------~----~------~--~---
Reply all
Reply to author
Forward
0 new messages
