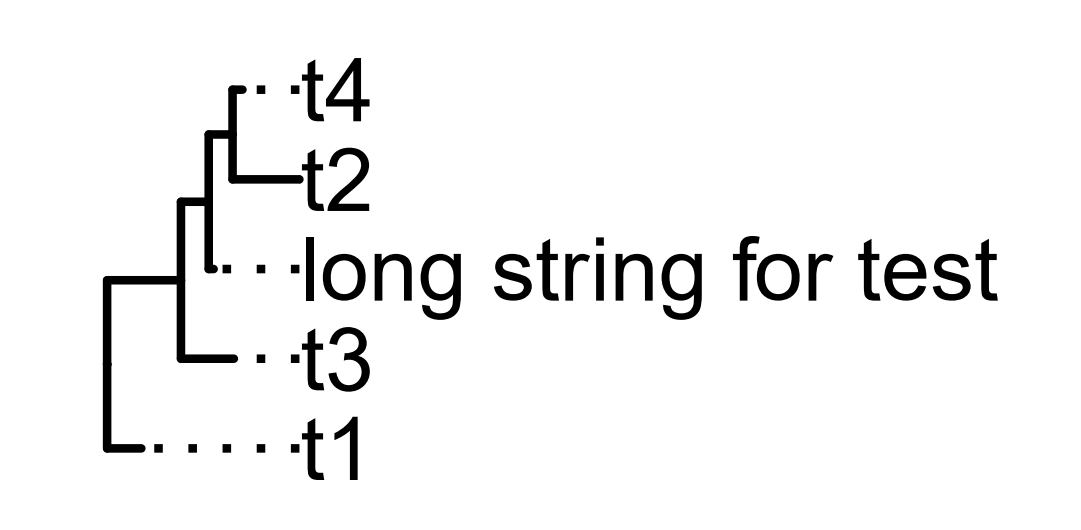align tip label to the end
1,849 views
Skip to first unread message
Pengcheng Yang
Sep 28, 2020, 4:23:29 AM9/28/20
to ggtree
Hi, there. Currently, the tip label is aligned to the tree. When the tree is vertically plot, the tip label is aligned to the left. Is it possible to align the tip label to the end? That is align to the right?
When the tip label is long, align-to-the-end will produce more space than align-to-the-tree.
xshua...@163.com
Sep 28, 2020, 5:24:36 AM9/28/20
to ggtree
Could you please provide a reproducible example?
Pengcheng Yang
Sep 28, 2020, 6:18:20 AM9/28/20
to ggtree
Hi,
Thank you for your reply.
The following code produced the tree illustrated below. I want to align the tip label to the right. Currently, they are aligned to the left.
set.seed(2015-12-21)
tree <- rtree(5)
tree$tip.label[2] <- "long string for test"
ggtree(tree) + xlim(0, 9) + geom_tiplab(align=TRUE)
Best,
Pengcheng
Pengcheng Yang
Sep 28, 2020, 10:23:58 PM9/28/20
to ggtree
The most difficult is it hard to compare the string width with the length of the dashed line. I have tried to add multiple "-" to the left of the labels, but the right end is not aligned well. Please help me.
Best,
Pengcheng
xshua...@163.com
Oct 7, 2020, 4:32:07 AM10/7/20
to ggtree
It might be not an universal feature. ggtree has not provided the method to do this. I tried to set `hjsut=1`,but the line and text are overlap. Or you consider to use 'long str\ning long string' to split long labels
Yonghe Xia
Oct 7, 2020, 10:53:10 AM10/7/20
to ggtree
Make all names of a character vector the same length is a considerable way.
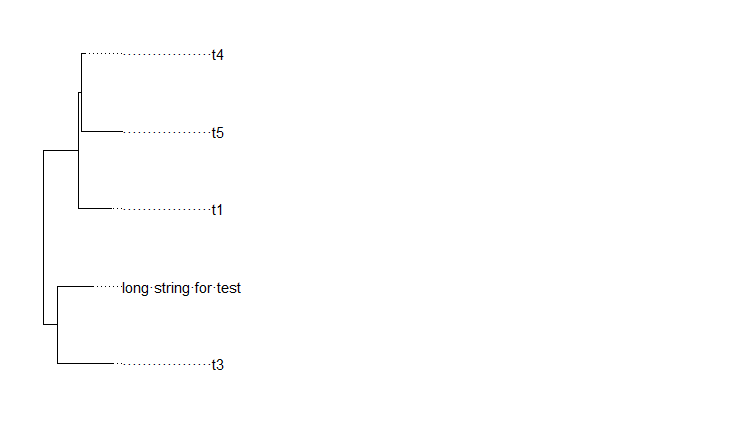

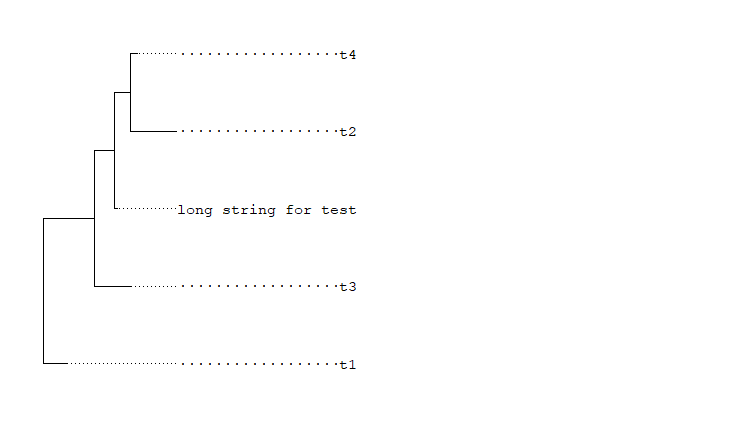
But there is a fact that the same length of strings doesn't equal the same length of labels displayed in the plot.
fonts also make an effect of length.
Here is an example for you to reference
```R
library(ggtree)
set.seed(2015-12-21)
tree <- rtree(5)
tree$tip.label[2] <- "long string for test"
newname <- gsub("\\s", "·", format(tree$tip.label, width=max(nchar(tree$tip.label)),justify = "right"))
tree$tip.label <- newname
# wrong font make lables not aligned
ggtree(tree) + xlim(0, 9) + geom_tiplab(align=TRUE)
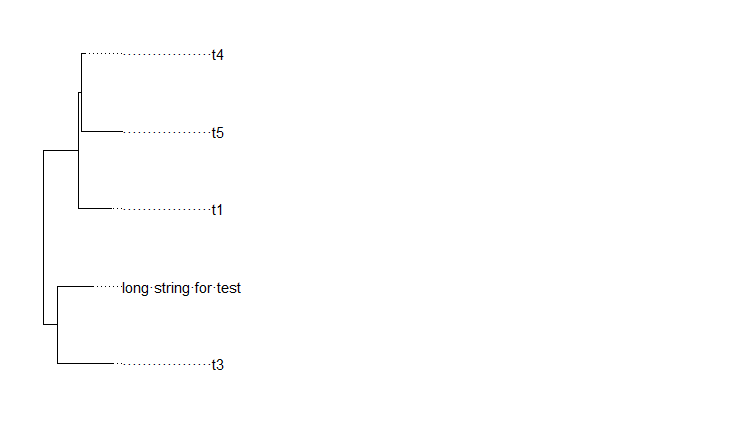
# Choose the right font
ggtree(tree) + xlim(0, 9) + geom_tiplab(align=TRUE,family="mono")

By the way, It makes the white spaces of the long name replaced when replace white spaces of all labels.
There is another method
```R
library(stringr)
newname <- str_pad(tree$tip.label, max(nchar(tree$tip.label)), side="left", pad="·")
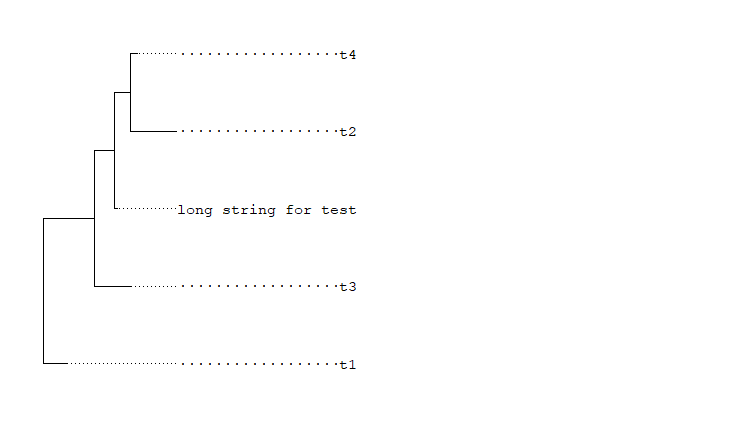
Pengcheng Yang
Oct 9, 2020, 2:56:03 AM10/9/20
to bioc-...@googlegroups.com
Hi Yonghe,
Thank you for your solution, which is practical for this problem.
Best,
Pengcheng
--
1. G Yu*. Using ggtree to visualize data on tree-like structures. Current Protocols in Bioinformatics. 2020, 69:e96. https://doi.org/10.1002/cpbi.96
2. LG Wang, TTY Lam, S Xu, Z Dai, L Zhou, T Feng, P Guo, CW Dunn, BR Jones, T Bradley, H Zhu, Y Guan, Y Jiang, G Yu*. treeio: an R package for phylogenetic tree input and output with richly annotated and associated data. Molecular Biology and Evolution. 2020, 37(2):599-603. http://dx.doi.org/10.1093/molbev/msz240
3. G Yu*, TTY Lam, H Zhu, Y Guan*. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Molecular Biology and Evolution, 2018, 35(2):3041-3043. https://doi.org/10.1093/molbev/msy194
4. G Yu, DK Smith, H Zhu, Y Guan, TTY Lam*. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data, Methods in Ecology and Evolution, 2017, 8(1):28-36. https://doi.org/10.1111/2041-210X.12628
5. Book: https://yulab-smu.top/treedata-book/
---
You received this message because you are subscribed to a topic in the Google Groups "ggtree" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/bioc-ggtree/INJ0Nfkq3b0/unsubscribe.
To unsubscribe from this group and all its topics, send an email to bioc-ggtree...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/bioc-ggtree/ffab3d5b-9b41-4291-8fc5-7afde2cf4870n%40googlegroups.com.
Yu, Guangchuang
Oct 10, 2020, 3:45:44 AM10/10/20
to Pengcheng Yang, ggtree
example presented in http://yulab-smu.top/treedata-book/chapter12.html#padding-taxa-labels.
You received this message because you are subscribed to the Google Groups "ggtree" group.
To unsubscribe from this group and stop receiving emails from it, send an email to bioc-ggtree...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/bioc-ggtree/39e5c7bd-756e-70cf-c9c6-6d020b4854b6%40gmail.com.
--
--~--~---------~--~----~------------~-------~--~----~
Guangchuang Yu PhD
Professor, Associate Director
Department of Bioinformatics
School of Basic Medical Sciences
Southern Medical University
Guangzhou, China
-~----------~----~----~----~------~----~------~--~---
Reply all
Reply to author
Forward
0 new messages