Debugging ChEBI and BFO2
21 views
Skip to first unread message
Alan Ruttenberg
May 14, 2013, 1:36:18 AM5/14/13
to Janna Hastings, bfo-owl-devel
I'm adding a test case that Janna Hasting sent me - classifying ChEBI with the mapping
chebi:has_part = has part at all times
chebi:has_role = bearer of at all times
chebi:role < realizable entity
chebi:biological role < function
chebi:application < chebi:role
chebi:chemical role < disposition
chebi:group < fiat object
chebi:atom < material entity
chebi:chemical_entity < material entity
chebi:subatomic_particle < material entity
chebi:chemical_substance < object
chebi:molecular entity < object
= is equivalentProperties
< is subClassOf
The issue that Janna raised was that importing BFO2 made the classification time much much longer, and doing the above mapping was worse.
Earlier in the day I tried classifying and found that it was very fast. I am redoing this now.
[Macbook Pro, i7 2.66Ghz, 8G ram, protege JVM -Xms1000M -Xmx4000M. After classification by Fact++ java uses about 1.2G. Reasoner settings are at protege defaults]
It is still fast and I find that there are many unsatisfiable classes. This seems to boil down to the domain and range restrictions on has part:

Because role is subclass of realizable entity, and the other two subclass of material entity, the domain and range are effectively owl:Nothing, as these are disjoint in BFO2. I suggest that ChEBI makes them disjoint as well. Not doing so is why ChEBI wasn't seeing the unsatisfiable classes.
Because of this, classification using BFO2 is actually substantially faster than it is without mapping to BFO2, because almost everything is classified under Nothing. However switching to the inferred view can be painfully slow because of all the classes stacked under Nothing.
The domain of has_role is the intersection of chemical entity, subatomic particle, and application. This too is empty as this amounts to the intersection of material entity and role.
Using the explanation facility we see, for example:

I've removed all the domain and range statements for has_part and has_role.
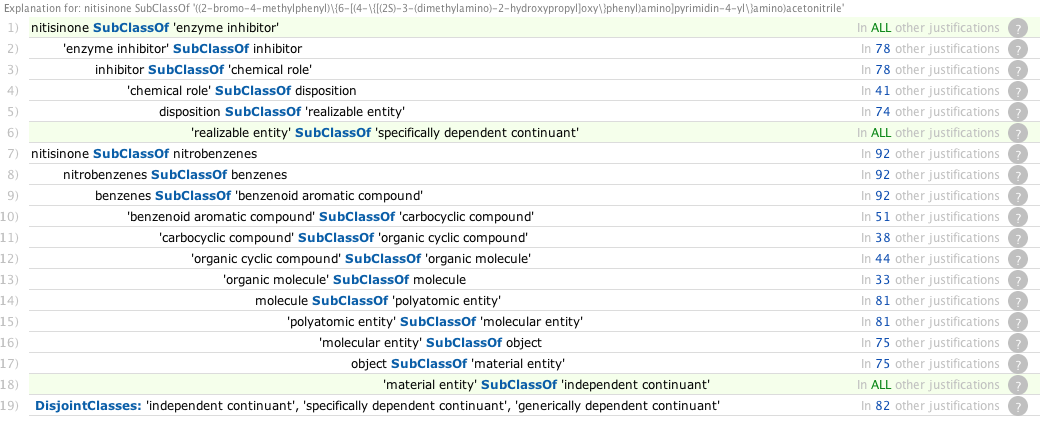
After doing so, classification slows down, taking perhaps 3 minutes, with ram usage up to about 1.5G. There remains a single unsatisfiable class: nitisinone, CHEBI_50378. From the time of display of clicking on the inferred classes tab, opening Nothing, and clicking the class in question, it takes another couple of minutes (I believe this can be improved by precomputing more using the reasoner settings).
Computing the explanations pushes memory usage up to 2.5G, with the following explanation, but the reason is apparent from the ontobee view http://purl.obolibrary.org/obo/CHEBI_50378 which shows the superclasses to be
The middle is a role while the others are material entities. These are disjoint in BFO2. The correction would be to change superclass enzyme inhibitor to has_role enzyme inhibitor
With this fix, the ontology is consistent.
Optimal settings for the task of reviewing ChEBI for inconsistencies would be to set the reasoner settings for display to be only unsatisfiability, or also superClasses in order to debug, with with either no precomputation (default, and fastest to classify) or precomputation of the class hierarchy, (might be reasonable compromise between speed of reasoning, speed of navigation in protege).
Classification of the ontology with the latter settings took 7.5 minutes, using 1.5G of RAM.
Files are in the repository at trunk/src/tests/chebi-classification/ - the top level there are the consistent files. The original files are in the subdirectory "as-submitted".
-Alan
-Alan
Alan Ruttenberg
May 14, 2013, 1:46:56 AM5/14/13
to bfo-ow...@googlegroups.com, Janna Hastings
One last fact - reasoning with inclusion of BFO2 took 1.5 minutes and 1.2G of RAM. So there is clearly a slowdown associated with the current version. I haven't yet investigated whether this can be sped up. OTOH I also haven't added needed axioms to Chebi - equivalent domain and range restrictions and disjoints - to see how long reasoning with these would be.
-Alan
dosumis
May 14, 2013, 4:25:10 AM5/14/13
to Alan Ruttenberg, Janna Hastings, bfo-owl-devel
Hi Alan,
Nice work. Would be good to send these to Ian Horrock's group to use as a test case for their combined elk/HermiT reasoner (MORe).
- David
On 14 May 2013, at 06:36, Alan Ruttenberg wrote:
I'm adding a test case that Janna Hasting sent me - classifying ChEBI with the mappingchebi:has_part = has part at all timeschebi:has_role = bearer of at all timeschebi:role < realizable entitychebi:biological role < functionchebi:application < chebi:rolechebi:chemical role < dispositionchebi:group < fiat objectchebi:atom < material entitychebi:chemical_entity < material entitychebi:subatomic_particle < material entitychebi:chemical_substance < objectchebi:molecular entity < object= is equivalentProperties< is subClassOfThe issue that Janna raised was that importing BFO2 made the classification time much much longer, and doing the above mapping was worse.Earlier in the day I tried classifying and found that it was very fast. I am redoing this now.[Macbook Pro, i7 2.66Ghz, 8G ram, protege JVM -Xms1000M -Xmx4000M. After classification by Fact++ java uses about 1.2G. Reasoner settings are at protege defaults]It is still fast and I find that there are many unsatisfiable classes. This seems to boil down to the domain and range restrictions on has part:
<image.png>
Because role is subclass of realizable entity, and the other two subclass of material entity, the domain and range are effectively owl:Nothing, as these are disjoint in BFO2. I suggest that ChEBI makes them disjoint as well. Not doing so is why ChEBI wasn't seeing the unsatisfiable classes.Because of this, classification using BFO2 is actually substantially faster than it is without mapping to BFO2, because almost everything is classified under Nothing. However switching to the inferred view can be painfully slow because of all the classes stacked under Nothing.The domain of has_role is the intersection of chemical entity, subatomic particle, and application. This too is empty as this amounts to the intersection of material entity and role.Using the explanation facility we see, for example:
<image.png>
I've removed all the domain and range statements for has_part and has_role.After doing so, classification slows down, taking perhaps 3 minutes, with ram usage up to about 1.5G. There remains a single unsatisfiable class: nitisinone, CHEBI_50378. From the time of display of clicking on the inferred classes tab, opening Nothing, and clicking the class in question, it takes another couple of minutes (I believe this can be improved by precomputing more using the reasoner settings).Computing the explanations pushes memory usage up to 2.5G, with the following explanation, but the reason is apparent from the ontobee view http://purl.obolibrary.org/obo/CHEBI_50378 which shows the superclasses to be
The middle is a role while the others are material entities. These are disjoint in BFO2. The correction would be to change superclass enzyme inhibitor to has_role enzyme inhibitor
<image.png>
With this fix, the ontology is consistent.Optimal settings for the task of reviewing ChEBI for inconsistencies would be to set the reasoner settings for display to be only unsatisfiability, or also superClasses in order to debug, with with either no precomputation (default, and fastest to classify) or precomputation of the class hierarchy, (might be reasonable compromise between speed of reasoning, speed of navigation in protege).Classification of the ontology with the latter settings took 7.5 minutes, using 1.5G of RAM.Files are in the repository at trunk/src/tests/chebi-classification/ - the top level there are the consistent files. The original files are in the subdirectory "as-submitted".
-Alan-Alan
--
You received this message because you are subscribed to the Google Groups "bfo-owl-devel" group.
To unsubscribe from this group and stop receiving emails from it, send an email to bfo-owl-deve...@googlegroups.com.
To post to this group, send email to bfo-ow...@googlegroups.com.
Visit this group at http://groups.google.com/group/bfo-owl-devel?hl=en-US.
For more options, visit https://groups.google.com/groups/opt_out.
Janna Hastings
May 14, 2013, 6:50:45 AM5/14/13
to Alan Ruttenberg, bfo-owl-devel
Great work Alan, thanks a lot!
The problem with nitisinone is precisely the sort of accident that we want to pick up with the use of the reasoner. We'll fix that.
We'll also fix the problem with the domain and range of the relations -- I can't even remember when those axioms were added but they are incorrect and aren't needed with a proper mapping to a version of BFO that includes relations.
Cheers, Janna
The problem with nitisinone is precisely the sort of accident that we want to pick up with the use of the reasoner. We'll fix that.
We'll also fix the problem with the domain and range of the relations -- I can't even remember when those axioms were added but they are incorrect and aren't needed with a proper mapping to a version of BFO that includes relations.
Cheers, Janna
Janna Hastings
May 14, 2013, 6:53:07 AM5/14/13
to dosumis, Alan Ruttenberg, bfo-owl-devel
David,
Yes, good suggestion. I also have other nastier (for reasoners) versions of ChEBI to send to Ian Horrock's group, with equivalence class axioms for many chemical entity classes. Some of them make use of cardinality which definitely poses a performance burden without, I think, being due to an error of the sort Alan has picked up here.
Cheers, JannaYes, good suggestion. I also have other nastier (for reasoners) versions of ChEBI to send to Ian Horrock's group, with equivalence class axioms for many chemical entity classes. Some of them make use of cardinality which definitely poses a performance burden without, I think, being due to an error of the sort Alan has picked up here.
Reply all
Reply to author
Forward
0 new messages
