Plot consensus tree with HPD intervals
46 views
Skip to first unread message
Emma Kopp
Feb 28, 2023, 12:42:54 PM2/28/23
to beast-users
Hi everyone,
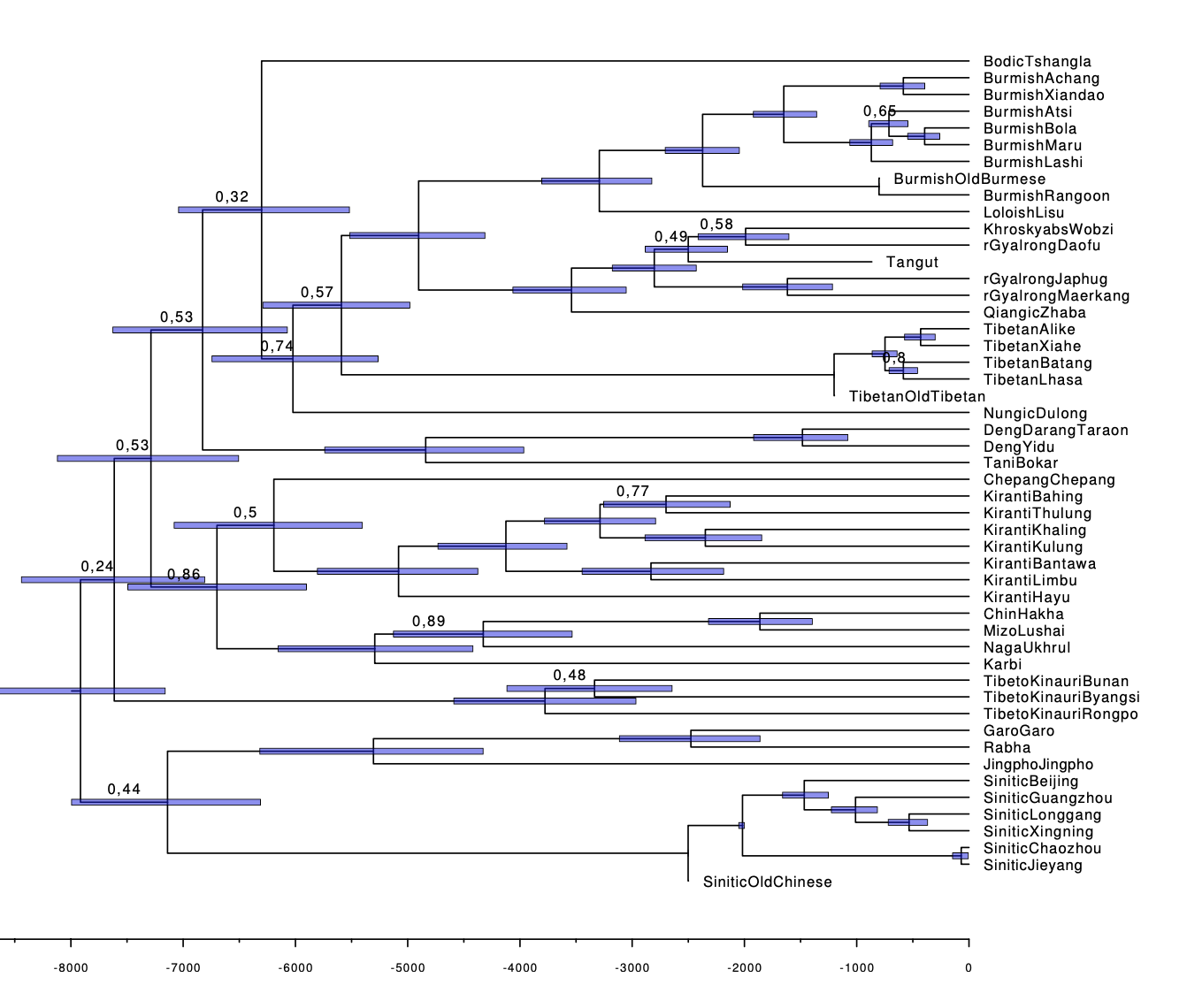
I would like to plot a consensus tree with the HPD intervals visibles as in attached photo.
I tried to :
1. Compute the consensus tree with r
2. Plot it with FigTree
I don't see any posterior probability values when I load a tree file in R. Otherwise, I read it's possible with FigTree to compute the posterior values/intervals on the NodeLabel level.
Thank you for your help,
Emma
ps : I use BEAST 2.6 and R to analyse the results

Jordan Douglas
Feb 28, 2023, 2:19:06 PM2/28/23
to beast-users
Hi Emma,
I'm not so familiar with the consensus tree method in R, but the only way FigTree can know the posterior values (or any other node metadata beyond branch length and label) is if the annotaions are being generated in the nexus/newick file. If you look at the nexus file in a text editor, you should be able to see what the R library is saving. A node with metadata should look like this (where the metdata is in bold):
1[&height=1.4553284949303256E-15,height_95%_HPD={0.0,2.6645352591003757E-15},height_median=1.7763568394002505E-15,height_range={0.0,3.552713678800501E-15},posterior=0.974232311]:1.9733814483511611
In general, R phylogenetic packages like ape tend to ignore this metadata [&...] which could be why the posterior support was lost. The Phylotate package for R may be of help here
If R doesn't behave then I suggest using the beast treeannotator application for generating the maximum clade credibility tree, which will directly load into fig tree with node HPDs and posterior support
Cheers,
Jordan
Emma Kopp
Feb 28, 2023, 5:22:49 PM2/28/23
to beast-users
Hello,
Thank you for your answer. Indeed for a maximum clade credibility tree, TreeAnnotator is perfect. But I also want to do it with consensus tree but it seems hard to compute a consensus tree and keep the incertitude values in the tree...
I am not sure that the phylotate package will help or maybe i didn't find the right function.
Thank you for your help,
Emma
Reply all
Reply to author
Forward
0 new messages
