Markers at the wrong end of a linkage group
68 views
Skip to first unread message
Etienne Dvorak
Nov 23, 2022, 8:15:01 AM11/23/22
to R/qtl discussion
Hello,
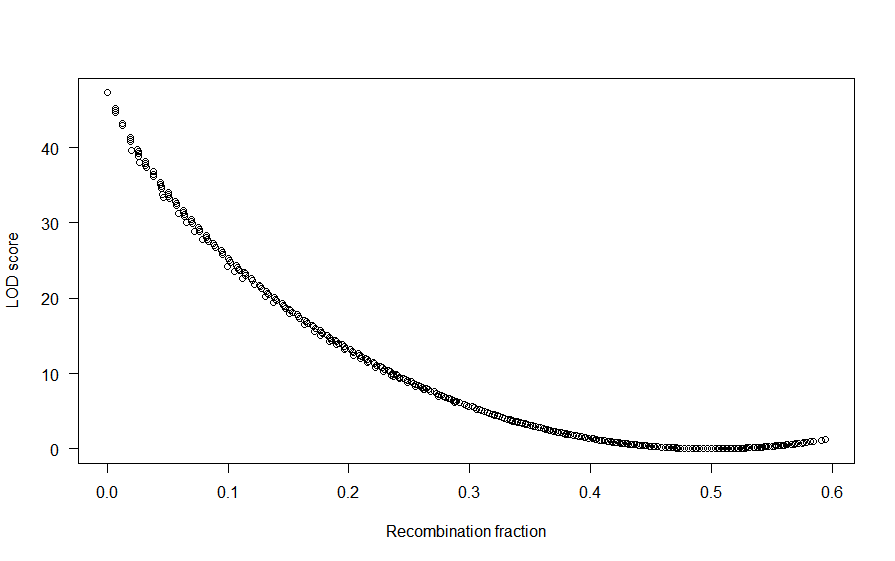
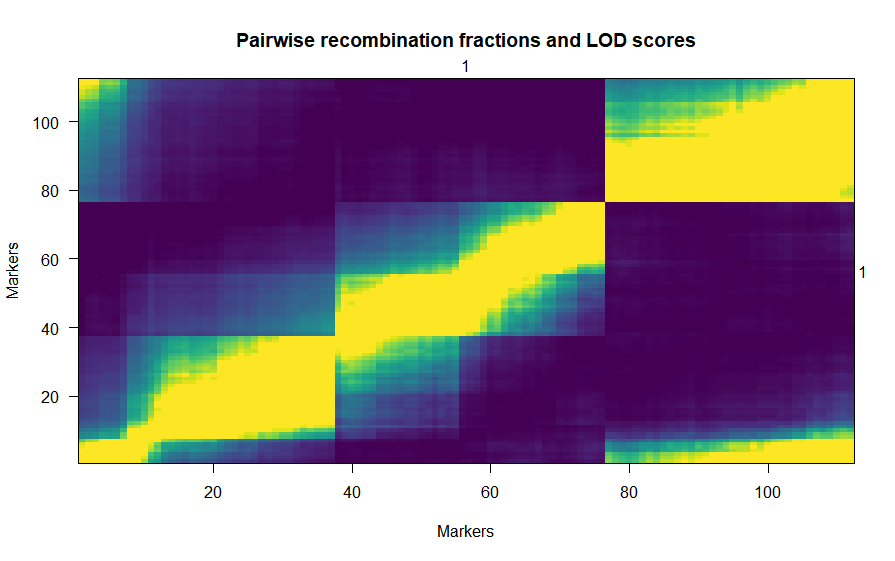
I am building a map using a backcross dataset of 1060 markers and 159 individuals. The package version is 1.56.
Here is my problem: there is a huge gap of ~100 cM in linkage group 1.
In fact, the group of markers after the gap is not linked at all to those before the gap... but they are linked to the other end of the linkage group!


Code used:
> rf = est.rf(testcross)
> LG = formLinkageGroups(rf, max.rf=0.35, min.lod=8) #same groups with min.lod=12
> LG = formLinkageGroups(rf, max.rf=0.35, min.lod=8,reorgMarkers=TRUE)
> map = orderMarkers(LG,map.function="kosambi",use.ripple = TRUE,error.prob=0.005)
I tried to manually change the marker order like this:
> compareorder(map,chr=1,c(78:112,1:77),error.prob=0.005,map.function="kosambi")
LOD length
orig 0.000 248.3662
new 35.856 158.1063
Warning message:
In est.map(cross, error.prob = error.prob, map.function = map.function, :
Didn't converge!
orig 0.000 248.3662
new 35.856 158.1063
Warning message:
In est.map(cross, error.prob = error.prob, map.function = map.function, :
Didn't converge!
Indeed it seems much better, and it is consistent with other linkage information that I have. The failure to converge might be a concern though?
What could cause this issue?
NB: I dropped redundant and distorted makers beforehand. I also removed some individuals that have abnormally high heterozygosity rate.
Karl Broman
Nov 23, 2022, 8:25:01 AM11/23/22
to R/qtl discussion
Yes, that block of markers at the end clearly belong at the beginning, as you found in compareorder().
the warning about "didn't converge" comes from trying to estimate the genetic map, which uses an iterative algorithm. But it might come from estimation with your original order, rather than estimation with the new order.
I'd use switch.order() to switch to the alternate order, and use est.map() to re-estimate the inter-marker distances. If you get the warning message about convergence, you can increase the maxit argument.
karl
Etienne Dvorak
Nov 29, 2022, 4:28:29 AM11/29/22
to R/qtl discussion
Thank you for your response.
The manual correction seems correct. I'm still confused about why orderMarkers consistently produces this gap (I tried to re-run the function several times and it always appeared).
Karl Broman
Nov 29, 2022, 5:08:49 PM11/29/22
to R/qtl discussion
orderMarkers() uses a pretty simplistic method. I don't think it's worth thinking too hard about why it's giving consistently bad results.
karl
Reply all
Reply to author
Forward
0 new messages
