Analysis of Control treatment in pre post variable
Manoj Diwakar
| Pre-test Scores | Posttest Scores |
| Control 84 | 115 |
| control 95 | 85 |
| control 103 | 102 |
| control 106 | 108 |
| control 101 | 100 |
| treatment 111 | 89 |
| treatment 104 | 106 |
| treatment 106 | 106 |
| treatment 108 | 99 |
Assistant Professor
Jawaharlal Nehru University, New Delhi-110067, India.
Marc Schwartz
You don't make it clear if the patients were prospectively randomized to treatment or not, but in general, the ANCOVA methodology described in the above has been shown to work in either scenario.
> DF
Group Pre Post
1 control 84 115
2 control 95 85
3 control 103 102
4 control 106 108
5 control 101 100
6 treatment 111 89
7 treatment 104 106
8 treatment 106 106
9 treatment 108 99
> anova(lm(Post ~ Pre * Group, data = DF))
Analysis of Variance Table
Response: Post
Df Sum Sq Mean Sq F value Pr(>F)
Pre 1 75.76 75.758 0.7764 0.4186
Group 1 8.88 8.883 0.0910 0.7750
Pre:Group 1 128.39 128.389 1.3158 0.3032
Residuals 5 487.86 97.572
Since the p value for the interaction term is 0.3032, it would be reasonable to consider dropping the term here, yielding the model:
> summary(lm(Post ~ Pre + Group, data = DF))
Call:
lm(formula = Post ~ Pre + Group, data = DF)
Residuals:
Min 1Q Median 3Q Max
-18.3424 -0.6404 2.4931 5.4007 9.9314
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 148.8894 54.7863 2.718 0.0348
Pre -0.4794 0.5583 -0.859 0.4234
Grouptreatment 2.5307 8.6053 0.294 0.7786
Residual standard error: 10.13 on 6 degrees of freedom
Multiple R-squared: 0.1208, Adjusted R-squared: -0.1723
F-statistic: 0.412 on 2 and 6 DF, p-value: 0.6797
So, in the above data, "control" is the reference level for treatment group. The value of 2.5307 for the beta coefficient for Group indicates that the treatment group's post value is 2.5307 points higher than the control group's post value, on average. That is your mean treatment effect in this sample. However, the p value for Group is 0.7786, which means that there is insufficient evidence to reject the null hypothesis of no difference between the groups.
Rich Ulrich
Sent: Friday, February 10, 2023 1:25 AM
To: meds...@googlegroups.com <meds...@googlegroups.com>
Subject: {MEDSTATS} Analysis of Control treatment in pre post variable
--
To post a new thread to MedStats, send email to MedS...@googlegroups.com .
MedStats' home page is http://groups.google.com/group/MedStats .
Rules: http://groups.google.com/group/MedStats/web/medstats-rules
---
You received this message because you are subscribed to the Google Groups "MedStats" group.
To unsubscribe from this group and stop receiving emails from it, send an email to medstats+u...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/medstats/CADTHf4tc5ZSjgRx81gBkc%3Dsf8nar7JAuMxb%2BnTNnH_cFGUoRuA%40mail.gmail.com.
Manoj Diwakar
summary (DF)
Group GpCont2Exp1 Class & section Gendar GendM1F2 PretestScores
Length:70 Min. :1.0 Length:70 Length:70 Min. :1.0 Min. : 70.00
Class :character 1st Qu.:1.0 Class :character Class :character 1st Qu.:1.0 1st Qu.: 90.25
Mode :character Median :1.5 Mode :character Mode :character Median :1.5 Median :101.00
Mean :1.5 Mean :1.5 Mean : 98.19
3rd Qu.:2.0 3rd Qu.:2.0 3rd Qu.:105.75
Max. :2.0 Max. :2.0 Max. :129.00
PosttestScores
Min. : 74.0
1st Qu.:100.0
Median :109.0
Mean :110.4
3rd Qu.:122.2
Max. :144.0 anova(lm(PosttestScores ~ PretestScores * GpCont2Exp1 , data = DF))
Analysis of Variance Table
Response: PosttestScores
Df Sum Sq Mean Sq F value Pr(>F)
PretestScores 1 512.5 512.5 4.0604 0.047976 *
GpCont2Exp1 1 8052.6 8052.6 63.7971 2.79e-11 ***
PretestScores:GpCont2Exp1 1 940.3 940.3 7.4496 0.008126 **
Residuals 66 8330.7 126.2
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
all:
lm(formula = PosttestScores ~ PretestScores + GpCont2Exp1, data = DF)
Residuals:
Min 1Q Median 3Q Max
-27.790 -8.301 1.155 7.012 23.164
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 114.1537 12.3457 9.246 1.39e-13 ***
PretestScores 0.2897 0.1201 2.413 0.0186 *
GpCont2Exp1 -21.4959 2.8178 -7.629 1.12e-10 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 11.76 on 67 degrees of freedom
Multiple R-squared: 0.4802, Adjusted R-squared: 0.4647
F-statistic: 30.95 on 2 and 67 DF, p-value: 3.021e-10
Assistant Professor
Jawaharlal Nehru University, New Delhi-110067, India.
Assistant Professor
Jawaharlal Nehru University, New Delhi-110067, India.
Marc Schwartz
Dear Prof. Marc Jii,Yes this one piece of data but here are 70 subjects each in control and treatment.I am running on R
summary (DF) Group GpCont2Exp1 Class & section Gendar GendM1F2 PretestScores Length:70 Min. :1.0 Length:70 Length:70 Min. :1.0 Min. : 70.00 Class :character 1st Qu.:1.0 Class :character Class :character 1st Qu.:1.0 1st Qu.: 90.25 Mode :character Median :1.5 Mode :character Mode :character Median :1.5 Median :101.00 Mean :1.5 Mean :1.5 Mean : 98.19 3rd Qu.:2.0 3rd Qu.:2.0 3rd Qu.:105.75 Max. :2.0 Max. :2.0 Max. :129.00 PosttestScores Min. : 74.0 1st Qu.:100.0 Median :109.0 Mean :110.4 3rd Qu.:122.2 Max. :144.0
anova(lm(PosttestScores ~ PretestScores * GpCont2Exp1 , data = DF))Analysis of Variance Table Response: PosttestScores Df Sum Sq Mean Sq F value Pr(>F) PretestScores 1 512.5 512.5 4.0604 0.047976 * GpCont2Exp1 1 8052.6 8052.6 63.7971 2.79e-11 *** PretestScores:GpCont2Exp1 1 940.3 940.3 7.4496 0.008126 ** Residuals 66 8330.7 126.2 --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1all: lm(formula = PosttestScores ~ PretestScores + GpCont2Exp1, data = DF) Residuals: Min 1Q Median 3Q Max -27.790 -8.301 1.155 7.012 23.164 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 114.1537 12.3457 9.246 1.39e-13 *** PretestScores 0.2897 0.1201 2.413 0.0186 * GpCont2Exp1 -21.4959 2.8178 -7.629 1.12e-10 *** --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 11.76 on 67 degrees of freedom Multiple R-squared: 0.4802, Adjusted R-squared: 0.4647 F-statistic: 30.95 on 2 and 67 DF, p-value: 3.021e-10
This is the output and comments on this ...
Best Regards,
Dr. Manoj Kumar Diwakar, M.Sc., M.Phil.,Ph.D. (Statistics)
Assistant Professor
Jawaharlal Nehru University, New Delhi-110067, India.Email id: Manojdiw...@gmail.com & mobile-09990346151Area of Specialisation: Statistics, Econometric and Applied MathematicsResearch Methodology -Quantitative Methods, Health Economics, Clinical Trial-Biostatistics
Data analysis and Software: SAS, SPSS, R
On Fri, Feb 10, 2023 at 6:45 PM Marc Schwartz <marc_s...@me.com> wrote:
--Best Regards,
Dr. Manoj Kumar Diwakar, M.Sc., M.Phil.,Ph.D. (Statistics)
Assistant Professor
Centre for Economic Studies & Planning (CESP), School of Social Sciences (SSS-II),
Jawaharlal Nehru University, New Delhi-110067, India.
Email id: Manojdiw...@gmail.com & mobile-09990346151Area of Specialisation: Statistics, Econometric and Applied MathematicsResearch Methodology -Quantitative Methods, Health Economics, Clinical Trial-BiostatisticsData analysis and Software: SAS, SPSS, R, STATA, SPSS AMOS
--
--
To post a new thread to MedStats, send email to MedS...@googlegroups.com .
MedStats' home page is http://groups.google.com/group/MedStats .
Rules: http://groups.google.com/group/MedStats/web/medstats-rules
---
You received this message because you are subscribed to the Google Groups "MedStats" group.
To unsubscribe from this group and stop receiving emails from it, send an email to medstats+u...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/medstats/CADTHf4tVCddOw%2BOXujgDXM6SadZ8PD8xC8Kr0zQKu1qq6_SeUg%40mail.gmail.com.
Karl Ove Hufthammer
Karl Ove Hufthammer
3. At least based upon the output from the ANOVA below, the p value for the interaction term is 0.008126. If a low p value (p < 0.05) is the case for the interaction term in the correct model on your full dataset, that means that you should retain the interaction term in your final model. Thus, you cannot interpret an overall main treatment effect in the presence of the interaction. You would need to generate predictions from the model, at varying values of interest over the range of the Pre Test Scores, the same values for each group, to have a sense for the magnitude and direction of the inter-group differences at each value.
One quick and easy way to do this in R, is to use the plot_model() function in the sjPlot package. Example:
mod = lm(PosttestScores ~
PretestScores * Group, data = DF)
library(sjPlot)
plot_model(mod,
type = "pred",
terms = c("PretestScores", "Group"))
You may also want to add the argument ‘show.data
= TRUE’, to overlay the original data on the prediction
plot.
-- Karl Ove Hufthammer
Manoj Diwakar
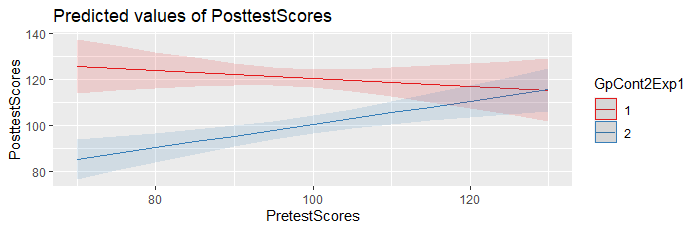
--
--
To post a new thread to MedStats, send email to MedS...@googlegroups.com .
MedStats' home page is http://groups.google.com/group/MedStats .
Rules: http://groups.google.com/group/MedStats/web/medstats-rules
---
You received this message because you are subscribed to the Google Groups "MedStats" group.
To unsubscribe from this group and stop receiving emails from it, send an email to medstats+u...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/medstats/82bd19b5-3a94-45ef-4630-b435d5440f5f%40huftis.org.
