BiocParallel errors when running crossvalidate()
51 views
Skip to first unread message
Dmitry Leontyev
Feb 9, 2022, 2:28:44 PM2/9/22
to Cardinal MSI Help
Hello,
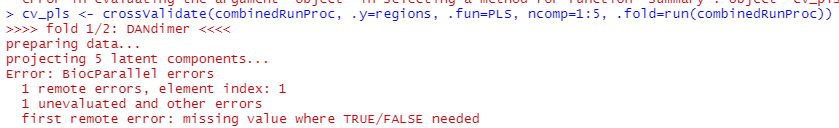
*Updated and deleted my previous post on this topic.*
I am try to call crossValidate() on my data set, but am getting the following error:

Where in my call to crossValidate is a TRUE/FALSE value missing? Code included below.
Thank you,
Dmitry
------Code------------
#defines paths
pathtest1 <- "C:/Users/Dima/Downloads/DANdimer.imzML"
pathtest2 <- "C:/Users/Dima/Downloads/LPC160.imzML"
#loads data
test1 <- readMSIData(pathtest1, mass.range=c(730,830), resolution=100, units="ppm", attach.only=TRUE)
test2 <- readMSIData(pathtest2, mass.range=c(730,830), resolution=100, units="ppm", attach.only=TRUE)
#sets centroided as TRUE, which is necessary for combination and combines runs
centroided(test1) <- TRUE
centroided(test2) <- TRUE
combinedRun <- Cardinal::combine(test1,test2)
#pre-process (in future might want to select ROIs before pre-process to reduce processing time)
combinedRunPre <- combinedRun %>% normalize(method="rms", delay=TRUE) %>% peakFilter(freq.min=0.01)
processingData(combinedRunPre)
combinedRunProc <- process(combinedRunPre,centroided=TRUE,plot=TRUE)
#ROI selection and subsetting and combining into regions
sampleA <- selectROI(combinedRunProc, mz=734.569, subset=run(combinedRunProc) == "DANdimer")
sampleB <- selectROI(combinedRunProc, mz=734.569, subset=run(combinedRunProc) == "LPC160")
regions <- makeFactor(DANdimer=sampleA, LPC160=sampleB)
#pls
cv_pls <- crossValidate(combinedRunProc, .y=regions, .fun=PLS, ncomp=1:5, .fold=run(combinedRunProc))
summary(cv_pls)
pathtest1 <- "C:/Users/Dima/Downloads/DANdimer.imzML"
pathtest2 <- "C:/Users/Dima/Downloads/LPC160.imzML"
#loads data
test1 <- readMSIData(pathtest1, mass.range=c(730,830), resolution=100, units="ppm", attach.only=TRUE)
test2 <- readMSIData(pathtest2, mass.range=c(730,830), resolution=100, units="ppm", attach.only=TRUE)
#sets centroided as TRUE, which is necessary for combination and combines runs
centroided(test1) <- TRUE
centroided(test2) <- TRUE
combinedRun <- Cardinal::combine(test1,test2)
#pre-process (in future might want to select ROIs before pre-process to reduce processing time)
combinedRunPre <- combinedRun %>% normalize(method="rms", delay=TRUE) %>% peakFilter(freq.min=0.01)
processingData(combinedRunPre)
combinedRunProc <- process(combinedRunPre,centroided=TRUE,plot=TRUE)
#ROI selection and subsetting and combining into regions
sampleA <- selectROI(combinedRunProc, mz=734.569, subset=run(combinedRunProc) == "DANdimer")
sampleB <- selectROI(combinedRunProc, mz=734.569, subset=run(combinedRunProc) == "LPC160")
regions <- makeFactor(DANdimer=sampleA, LPC160=sampleB)
#pls
cv_pls <- crossValidate(combinedRunProc, .y=regions, .fun=PLS, ncomp=1:5, .fold=run(combinedRunProc))
summary(cv_pls)
-----------------------------------------
Reply all
Reply to author
Forward
0 new messages
