Importing data: stack mirror error
147 views
Skip to first unread message
Marcel Sayre
Aug 24, 2019, 2:53:10 PM8/24/19
to CATMAID
Hi all,
I've managed to get catmaid running on a development server using Nginx on Ubuntu 18, but I'm now having difficulty actually visualizing my image stack. I apologize if this has been discussed elsewhere, I went through previous related threads but am still having difficulty.
I am trying to import a data set of 10 image slices (greyscale, BF-SEM data) which have been exported as .jpg using the TrakEM2 "export for catmaid" option. The image data is in the following directory/hierarchy:
/home/user/catmaid/httpdocs/data/project1
/stack1
/project.yaml
Here is my project.yaml file (no tabs were used, only spaces):
Regarding permissions, running namei -l /home/user/catmaid/httpdocs/data/project1/stack1 returns the following:
f: /home/user/catmaid/httpdocs/data/project1/stack1
drwxr-xr-x root root /
drwxr-xr-x root root home
drwxrwxrwx user user user
drwxrwxrwx user user catmaid
drwxrwxrwx user user httpdocs
drwxrwxrwx user user data
drwxr-xr-x user user project1
drwxr-xr-x user user stack1
drwxr-xr-x root root /
drwxr-xr-x root root home
drwxrwxrwx user user user
drwxrwxrwx user user catmaid
drwxrwxrwx user user httpdocs
drwxrwxrwx user user data
drwxr-xr-x user user project1
drwxr-xr-x user user stack1
I uploaded the data using the admin "Import projects and image stacks". When I do so, I receive the following error in red:
In my settings.py file I have the following set:
CATMAID_IMPORT_PATH = '/home/user/catmaid/httpdocs/data'
IMPORTER_DEFAULT_IMAGE_BASE = 'http://localhost/catmaid/data'
IMPORTER_DEFAULT_IMAGE_BASE = 'http://localhost/catmaid/data'
Here's what my homepage looks like:
Lastly, I've attached my settings.py, configuration.py, and nginx.conf (from /etc/nginx/conf.d) here. Any help would be greatly appreciated! Also, I'm very new to all of this, so please dumb-down explanations if possible, and let me know if I can provide any other useful info! Thanks so much for any advice.
youmin Wu
Aug 24, 2019, 9:18:35 PM8/24/19
to CATMAID
I had the same problem for importing project. And also waiting someone who can save me. ^_^
Tom Kazimiers
Aug 29, 2019, 10:28:33 AM8/29/19
to cat...@googlegroups.com
Hi Marcel,
I believe the problem you run into is that you are missing an Nginx location
block that map the IMPORTER_DEFAULT_IMAGE_BASE URL to your CATMAID_IMPORT_PATH.
See below for more details.
On Sat, Aug 24, 2019 at 11:53:10AM -0700, Marcel Sayre wrote:
>I've managed to get catmaid running on a development server using Nginx
>on Ubuntu 18, but I'm now having difficulty actually visualizing my image
>stack. I apologize if this has been discussed elsewhere, I went through
>previous related threads but am still having difficulty.
Great that you managed to set up CATMAID using Nginx. What WSGI server
are you using? For testing the "manage.py runserver" is okay, but it is
not suited for production use. We have made good experience with uWSGI.
>I am trying to import a data set of 10 image slices (greyscale, BF-SEM
>data) which have been exported as .jpg using the TrakEM2 "export for
>catmaid" option. The image data is in the following directory/hierarchy:
>/home/user/catmaid/httpdocs/data/project1
> /stack1
> /project.yaml
This is a layout CATMAID should understand, yes.
>Here is my project.yaml file (no tabs were used, only spaces):
>
>[project.yaml screenshot]
It fits your above folder structure and should work.
>Regarding permissions, running *namei -l
>/home/user/catmaid/httpdocs/data/project1/stack1* returns the following:
make your image data accessible through Nginx. You can even be more
restrictive, because you probably don't want all folders between
/home/user and you data folder to be writable by all all group members
and all other users.
>I uploaded the data using the admin "Import projects and image stacks".
>When I do so, I receive the following error in red:
> [import confirmation view screenshot]
The importer tries to access the stack's so called canary tile, which is
used to indicate whether a stack is accessible or not. This is the stack
coordinate (0,0,0) by default.
As you seem to already know, you can finish the import regardless if you
want and fix this problem directly in the respective stack mirror entry.
A stack mirror contains the URL to your files for a particular stack. A
stack can have multiple mirrors, in your case it should be only one
though.
>In my settings.py file I have the following set:
>CATMAID_IMPORT_PATH = '/home/user/catmaid/httpdocs/data'
>IMPORTER_DEFAULT_IMAGE_BASE = 'http://localhost/catmaid/data'
These look okay it basically says you have to tell Nginx to map
'http://localhost/catmaid/data' to '/home/user/catmaid/httpdocs/data'.
It also matches the import confirmation view screenshot.
>Here's what my homepage looks like:
>
> [CATMAID front page screenshot]
Okay, great that you can load the front-page with all static files
correctly already. It also looks like you saved the import configuration
regardless of the unreachable canary tiles. Since the import looked
correct in general, it is likely that you don't need to change anything
in your CATMAID stacks and stack mirrors for now. The white space right
next to "project" should contain a sample image from your dataset.
CATMAID will look for a file called small.jpg for each section. Do you
happen to see them in your TrakEM2 export? Presumably, the sample image
not being visible has likely the same reason why the canary tiles were
not accessible.
Just in case you aren't aware of this, you can have different front
pages (called "data views" in CATMAID). By default there are only two
configured and you can change between them through the menu that appears
when you hover the mouse over the "Home" link in the upper left corner.
The version without images should give you a clearer picture of the
setup and allow you to try to open the stack regardless.
>Lastly, I've attached my settings.py, configuration.py, and nginx.conf
>(from /etc/nginx/conf.d) here. Any help would be greatly appreciated! Also,
>I'm very new to all of this, so please dumb-down explanations if possible,
>and let me know if I can provide any other useful info! Thanks so much for
>any advice.
Thanks for attaching all the relevant files. After looking at your Nginx
configuration, I believe the problem you run into is that you don't map
the IMPORTER_DEFAULT_IMAGE_BASE URL to the local CATMAID_IMPORT_PATH. To
do this, you could add a location block like this:
location /catmaid/data/ {
alias /home/user/catmaid/httpdocs/data/;
}
Does this help? If not, I'd be curious what the Nginx error.log file has
to say about access attempts to the tiles? I just browsed through our
data import documentation page [1] and couldn't find any mention of this
and I'll add a brief remark about required changes to the webserver
configuration to make the image data accessible in the first place.
Also, how comes you have to "proxy_pass" blocks (one for / and one for
/catmaid)?
Best,
Tom
I believe the problem you run into is that you are missing an Nginx location
block that map the IMPORTER_DEFAULT_IMAGE_BASE URL to your CATMAID_IMPORT_PATH.
See below for more details.
On Sat, Aug 24, 2019 at 11:53:10AM -0700, Marcel Sayre wrote:
>I've managed to get catmaid running on a development server using Nginx
>on Ubuntu 18, but I'm now having difficulty actually visualizing my image
>stack. I apologize if this has been discussed elsewhere, I went through
>previous related threads but am still having difficulty.
are you using? For testing the "manage.py runserver" is okay, but it is
not suited for production use. We have made good experience with uWSGI.
>I am trying to import a data set of 10 image slices (greyscale, BF-SEM
>data) which have been exported as .jpg using the TrakEM2 "export for
>catmaid" option. The image data is in the following directory/hierarchy:
>/home/user/catmaid/httpdocs/data/project1
> /stack1
> /project.yaml
>Here is my project.yaml file (no tabs were used, only spaces):
>
It fits your above folder structure and should work.
>Regarding permissions, running *namei -l
>/home/user/catmaid/httpdocs/data/project1/stack1* returns the following:
>
>f: /home/user/catmaid/httpdocs/data/project1/stack1
>drwxr-xr-x root root /
>drwxr-xr-x root root home
>drwxrwxrwx user user user
>drwxrwxrwx user user catmaid
>drwxrwxrwx user user httpdocs
>drwxrwxrwx user user data
>drwxr-xr-x user user project1
>drwxr-xr-x user user stack1
Thanks, that's useful information. The permissions should definitely
>f: /home/user/catmaid/httpdocs/data/project1/stack1
>drwxr-xr-x root root /
>drwxr-xr-x root root home
>drwxrwxrwx user user user
>drwxrwxrwx user user catmaid
>drwxrwxrwx user user httpdocs
>drwxrwxrwx user user data
>drwxr-xr-x user user project1
>drwxr-xr-x user user stack1
make your image data accessible through Nginx. You can even be more
restrictive, because you probably don't want all folders between
/home/user and you data folder to be writable by all all group members
and all other users.
>I uploaded the data using the admin "Import projects and image stacks".
>When I do so, I receive the following error in red:
The importer tries to access the stack's so called canary tile, which is
used to indicate whether a stack is accessible or not. This is the stack
coordinate (0,0,0) by default.
As you seem to already know, you can finish the import regardless if you
want and fix this problem directly in the respective stack mirror entry.
A stack mirror contains the URL to your files for a particular stack. A
stack can have multiple mirrors, in your case it should be only one
though.
>In my settings.py file I have the following set:
>CATMAID_IMPORT_PATH = '/home/user/catmaid/httpdocs/data'
>IMPORTER_DEFAULT_IMAGE_BASE = 'http://localhost/catmaid/data'
'http://localhost/catmaid/data' to '/home/user/catmaid/httpdocs/data'.
It also matches the import confirmation view screenshot.
>Here's what my homepage looks like:
>
Okay, great that you can load the front-page with all static files
correctly already. It also looks like you saved the import configuration
regardless of the unreachable canary tiles. Since the import looked
correct in general, it is likely that you don't need to change anything
in your CATMAID stacks and stack mirrors for now. The white space right
next to "project" should contain a sample image from your dataset.
CATMAID will look for a file called small.jpg for each section. Do you
happen to see them in your TrakEM2 export? Presumably, the sample image
not being visible has likely the same reason why the canary tiles were
not accessible.
Just in case you aren't aware of this, you can have different front
pages (called "data views" in CATMAID). By default there are only two
configured and you can change between them through the menu that appears
when you hover the mouse over the "Home" link in the upper left corner.
The version without images should give you a clearer picture of the
setup and allow you to try to open the stack regardless.
>Lastly, I've attached my settings.py, configuration.py, and nginx.conf
>(from /etc/nginx/conf.d) here. Any help would be greatly appreciated! Also,
>I'm very new to all of this, so please dumb-down explanations if possible,
>and let me know if I can provide any other useful info! Thanks so much for
>any advice.
configuration, I believe the problem you run into is that you don't map
the IMPORTER_DEFAULT_IMAGE_BASE URL to the local CATMAID_IMPORT_PATH. To
do this, you could add a location block like this:
location /catmaid/data/ {
alias /home/user/catmaid/httpdocs/data/;
}
Does this help? If not, I'd be curious what the Nginx error.log file has
to say about access attempts to the tiles? I just browsed through our
data import documentation page [1] and couldn't find any mention of this
and I'll add a brief remark about required changes to the webserver
configuration to make the image data accessible in the first place.
Also, how comes you have to "proxy_pass" blocks (one for / and one for
/catmaid)?
Best,
Tom
youmin Wu
Aug 30, 2019, 7:11:01 AM8/30/19
to CATMAID
Hi Tom and Marcel,
I've solved this problem.
Besides making the right config following Tom's instruction, I also need to enable the nginx folder access permission in nginx.conf, by setting "autoindex on" under "server" block.
Tom Kazimiers
Aug 30, 2019, 9:24:29 AM8/30/19
to cat...@googlegroups.com
Hi Youmin,
On Fri, Aug 30, 2019 at 04:11:01AM -0700, youmin Wu wrote:
>I've solved this problem.
Great to hear you could find a solution! This sounds like you are now
able to view all your tiled data in CATMAID?
>Besides making the right config following Tom's instruction, I also
>need to enable the nginx folder access permission in nginx.conf, by
>setting "autoindex on" under "server" block.
Thanks for the additional info! What would help autoindex with though?
As far as I know this only allows Nginx to do a folder listing,
something that I have disabled on basically all setups I work with. This
setting shouldn't be needed for regular tile/data access. The CATMAID
front-end knows all needed URLs and won't ever request a directory
listing.
If you disable autoindex again, what exactly stops working for you?
Also, keep in mind that this setting is security relevant. It usually
discloses more information than necessary and you have to make sure that
you don't store any sensitive data in a publicly accessible folder,
especially with directory listing enabled: For an attacker it seems much
quicker to scan such a listing for potentially "useful" sounding files
(e.g. backups, config files with DB passwords, etc.) compared to
randomly requesting file names (without knowing the listing, which still
happens a lot on production systems). It's probably fine though if you
enable this for the image data, but I would try to find a configuration
that doesn't need it.
Best,
Tom
On Fri, Aug 30, 2019 at 04:11:01AM -0700, youmin Wu wrote:
>I've solved this problem.
able to view all your tiled data in CATMAID?
>Besides making the right config following Tom's instruction, I also
>need to enable the nginx folder access permission in nginx.conf, by
>setting "autoindex on" under "server" block.
As far as I know this only allows Nginx to do a folder listing,
something that I have disabled on basically all setups I work with. This
setting shouldn't be needed for regular tile/data access. The CATMAID
front-end knows all needed URLs and won't ever request a directory
listing.
If you disable autoindex again, what exactly stops working for you?
Also, keep in mind that this setting is security relevant. It usually
discloses more information than necessary and you have to make sure that
you don't store any sensitive data in a publicly accessible folder,
especially with directory listing enabled: For an attacker it seems much
quicker to scan such a listing for potentially "useful" sounding files
(e.g. backups, config files with DB passwords, etc.) compared to
randomly requesting file names (without knowing the listing, which still
happens a lot on production systems). It's probably fine though if you
enable this for the image data, but I would try to find a configuration
that doesn't need it.
Best,
Tom
youmin Wu
Aug 31, 2019, 3:17:24 AM8/31/19
to CATMAID
Hi Tom,
I remove the nginx folder access permission and reimport the project. You are right CATMAID don't need this permission, the CATMAID read the images correctly, except that CATMAID can't find the small.jpg.
But the "Stack Image Base" always shown in red.
youmin Wu
Aug 31, 2019, 3:42:40 AM8/31/19
to CATMAID
Hi Tom,
![]()
![]()
I have another question.
As shown below, I can load the image in the web.
But I can't find those tools in the web browser which introduced in CATMAID's document.
Do you know how I can use these tools? Thank you very much.
Tom Kazimiers
Aug 31, 2019, 8:35:19 PM8/31/19
to cat...@googlegroups.com
Hi Youmin,
It looks like there were no images attached to your emails.
On Sat, Aug 31, 2019 at 12:42:39AM -0700, youmin Wu wrote:
>But I can't find those tools in the web browser which introduced in
>CATMAID's document.
>
toolbar. The visible tools are specified per user and you can change the
visibility settings for each tool in the each user's userprofile, which
you can edit in the /admin view. To do so, just edit a user and find the
tool configuration at the bottom of the page.
Also you can override the following default attributes in your
settings.py file and new users will start with these settings:
PROFILE_SHOW_TEXT_LABEL_TOOL = False
PROFILE_SHOW_TAGGING_TOOL = False
PROFILE_SHOW_CROPPING_TOOL = False
PROFILE_SHOW_TRACING_TOOL = False
PROFILE_SHOW_ONTOLOGY_TOOL = False
PROFILE_SHOW_ROI_TOOL = False
(As defined in `settings_base.py`.)
This is also explained in the documentation [1].
I have also update the configuration.py.example file and the
`create_configuration.py` script in the dev branch (next release) to
expose this configuration option also there by defining a
`catmaid_default_enabled_tools` attribute, which by default is defined
as `['tracing']`, i.e. the tracing tool is available by default for new
users. This list can hold the following options: 'cropping', 'tagging',
'textlabel', 'tracing', 'ontology' and 'roi'. If the attribute isn't
available, no change is made to the default tool options.
For this to take effect you have to run `create_configuration.py` again
though. In you case, you should be fine by just adding the above
PROFILE… lines to your settings.py and set the one to `True` for the
tools you'd like to see by default for new users.
The default tool options are specified in most setups, so I figured it
is reasonable to also expose it in the more top-level configuration.py.
Plus the tracing tool is something that is enabled by default in many
setups anyway
Best,
Tom
[1] https://catmaid.readthedocs.io/en/stable/userprofiles.html#user-profiles
It looks like there were no images attached to your emails.
On Sat, Aug 31, 2019 at 12:42:39AM -0700, youmin Wu wrote:
>But I can't find those tools in the web browser which introduced in
>CATMAID's document.
>
>Do you know how I can use these tools? Thank you very much.
I assume you are referring to the icons that are available from the top
toolbar. The visible tools are specified per user and you can change the
visibility settings for each tool in the each user's userprofile, which
you can edit in the /admin view. To do so, just edit a user and find the
tool configuration at the bottom of the page.
Also you can override the following default attributes in your
settings.py file and new users will start with these settings:
PROFILE_SHOW_TEXT_LABEL_TOOL = False
PROFILE_SHOW_TAGGING_TOOL = False
PROFILE_SHOW_CROPPING_TOOL = False
PROFILE_SHOW_TRACING_TOOL = False
PROFILE_SHOW_ONTOLOGY_TOOL = False
PROFILE_SHOW_ROI_TOOL = False
(As defined in `settings_base.py`.)
This is also explained in the documentation [1].
I have also update the configuration.py.example file and the
`create_configuration.py` script in the dev branch (next release) to
expose this configuration option also there by defining a
`catmaid_default_enabled_tools` attribute, which by default is defined
as `['tracing']`, i.e. the tracing tool is available by default for new
users. This list can hold the following options: 'cropping', 'tagging',
'textlabel', 'tracing', 'ontology' and 'roi'. If the attribute isn't
available, no change is made to the default tool options.
For this to take effect you have to run `create_configuration.py` again
though. In you case, you should be fine by just adding the above
PROFILE… lines to your settings.py and set the one to `True` for the
tools you'd like to see by default for new users.
The default tool options are specified in most setups, so I figured it
is reasonable to also expose it in the more top-level configuration.py.
Plus the tracing tool is something that is enabled by default in many
setups anyway
Best,
Tom
[1] https://catmaid.readthedocs.io/en/stable/userprofiles.html#user-profiles
youmin Wu
Aug 31, 2019, 9:58:39 PM8/31/19
to CATMAID
Hi Tom,
Yes! Those tools are exactly what I want.
Thank you very much!
Message has been deleted
youmin Wu
Aug 31, 2019, 10:54:26 PM8/31/19
to CATMAID
Hi Tom,
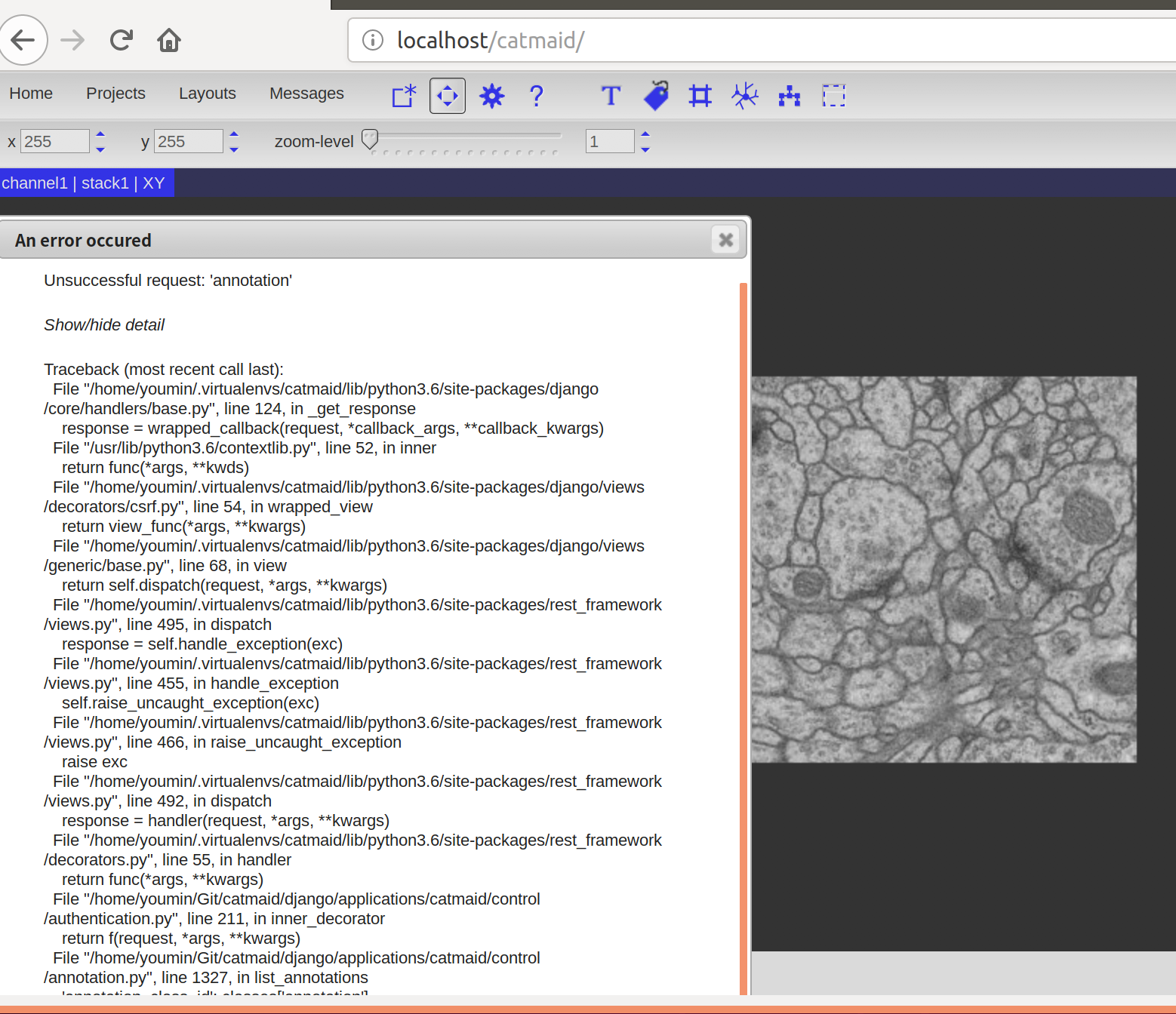
Could you please give me another help? ^_^
I loaded all the tools to the web page.
But I got an exception window when I enter the image page.
I didn't change any source code.

On Sunday, September 1, 2019 at 8:35:19 AM UTC+8, Tom Kazimiers wrote:
Tom Kazimiers
Aug 31, 2019, 11:14:11 PM8/31/19
to cat...@googlegroups.com
Hi Youmin,
On Sat, Aug 31, 2019 at 07:54:25PM -0700, youmin Wu wrote:
>Could you please give me another help? ^_^
>
>I loaded all the tools to the web page.
>But I got an exception window when I enter the image page.
>I didn't change any source code.
>[image: exception.png]
Would you mind copying the whole detailed exception text in the error
dialog and send it here?
And did this error shop up just when opening a stack or on another
action (e.g. selecting a tool from the toolbar)?
Best,
Tom
On Sat, Aug 31, 2019 at 07:54:25PM -0700, youmin Wu wrote:
>Could you please give me another help? ^_^
>
>I loaded all the tools to the web page.
>But I got an exception window when I enter the image page.
>I didn't change any source code.
>[image: exception.png]
dialog and send it here?
And did this error shop up just when opening a stack or on another
action (e.g. selecting a tool from the toolbar)?
Best,
Tom
Marcel Sayre
Sep 1, 2019, 2:20:31 PM9/1/19
to CATMAID
Hello Tom,
Thank you so much for that info! Inserting the Nginx block into the config file solved the problem and I'm now able to see and navigate my stack.
Great that you managed to set up CATMAID using Nginx. What WSGI server
are you using? For testing the "manage.py runserver" is okay, but it is
not suited for production use. We have made good experience with uWSGI.
I'm just running it using the development server at the moment, but the goal is to get it into production using uWSGI.
Thanks, that's useful information. The permissions should definitely
make your image data accessible through Nginx. You can even be more
restrictive, because you probably don't want all folders between
/home/user and you data folder to be writable by all all group members
and all other users.
Thanks for that info!
The white space right next to "project" should contain a sample image from your dataset.
CATMAID will look for a file called small.jpg for each section. Do you
happen to see them in your TrakEM2 export? Presumably, the sample image
not being visible has likely the same reason why the canary tiles were
not accessible.
Even though I am now able to see the data set, there is still no home page image. I checked the files and there are no small.jpg images.
Also, I had to use file type number 4 (file-based zoom directories) to get the stack mirror to recognize my stack. This didn't happen for the
first stack I tried, but did for the second (the first stack I tried had a small.jpg file and was exported in a way accessible using file type 1).
I'm fairly certain that I exported both stacks using the same process in CATMAID so I'm not entirely sure why they were organized differently.
Also, how comes you have to "proxy_pass" blocks (one for / and one for
/catmaid)?
To be completely honest, I have no idea! I had a friend of a friend help me set up the Nginx block, and I am unable to reach out to him
to ask why those both exist. However, when I try and remove the second proxy pass (for /), the page no longer works. Any idea why
this might be? (I can send additional error log info).
Up until now, our group has traced neurons using the skeletonize tool in Amira 5.3. My goal is to transfer all of our data from Amira
(stacks plus skeletons) into CATMAID to make CATMAID our primary tracing software. Now that I am able to navigate
an image stack, I've tried uploading some of the neuron skeletons we've traced thus far and I'm having an issue with coordinate values (I
exported the skeletons from Amira as SWC files and imported them into CATMAID using the importer widget). The skeletons look good in the
3D viewer, but appear to be in the wrong z-location (they appear ~ in the right x-y location, but I won't be able to tell for sure until the z is adjusted).
In our version of Amira, we've used um as the unit of measure (and there doesn't appear to be a preference setting to change unit values). So our stack,
having a resolution of 50 nm x 50 nm x 50 nm, was imported into Amira with a voxel size of 0.05 um^3. In CATMAID, I've tried changing the stack resolution
to both 50 nm^3 (which would be accurate for CATMAID's nm coordinate field) and 0.05 nm^3 for testing purposes. The skeleton appears off in both instances
although is much closer when imported using the 0.05 nm^3 value for the stack.
Using Amira, I can get the "bounding box" coordinates of the neuron, and convert the values to nm, but I'm not entirely sure how this would work.
For the stack itself, I converted the .am Amira file into .tiff using FIJI, and exported the stack for CATMAID using TrakEM2 (side note: I tried using
the "Amira as TrakEM2" import option, but I receive an error when I select my stack).
Do you have any advice for adjusting coordinate perimeters of neuron skeletons imported as SWC files? Does anyone have experience migrating
a project full of skeletons and stacks from Amira (or other platform) into CATMAID? Maybe I should post this as an entirely new thread?
Thanks again for your time, Tom!
warm regards,
Marcel
Tom Kazimiers
Sep 1, 2019, 7:35:50 PM9/1/19
to cat...@googlegroups.com
Hi Youmin,
On Sat, Aug 31, 2019 at 11:14:07PM -0400, Tom Kazimiers wrote:
>On Sat, Aug 31, 2019 at 07:54:25PM -0700, youmin Wu wrote:
>>Could you please give me another help? ^_^
>>
>>I loaded all the tools to the web page.
>>But I got an exception window when I enter the image page.
>>I didn't change any source code.
>>[image: exception.png]
>
>Would you mind copying the whole detailed exception text in the error
>dialog and send it here?
Sorry, I don't think I need the full stacktrace anymore. I pushed a
change to CATMAID's master branch that should hopefully fix the error
you see. Please try updating you local setup with this.
>And did this error shop up just when opening a stack or on another
>action (e.g. selecting a tool from the toolbar)?
I would still be curious though what exactly triggered this error.
Best,
Tom
On Sat, Aug 31, 2019 at 11:14:07PM -0400, Tom Kazimiers wrote:
>On Sat, Aug 31, 2019 at 07:54:25PM -0700, youmin Wu wrote:
>>Could you please give me another help? ^_^
>>
>>I loaded all the tools to the web page.
>>But I got an exception window when I enter the image page.
>>I didn't change any source code.
>>[image: exception.png]
>
>Would you mind copying the whole detailed exception text in the error
>dialog and send it here?
change to CATMAID's master branch that should hopefully fix the error
you see. Please try updating you local setup with this.
>And did this error shop up just when opening a stack or on another
>action (e.g. selecting a tool from the toolbar)?
Best,
Tom
Sanja Jasek
Sep 2, 2019, 7:10:26 AM9/2/19
to CATMAID
Hi Marcel,
On Sunday, 1 September 2019 19:20:31 UTC+1, Marcel Sayre wrote:
Also, how comes you have to "proxy_pass" blocks (one for / and one for
/catmaid)?To be completely honest, I have no idea! I had a friend of a friend help me set up the Nginx block, and I am unable to reach out to himto ask why those both exist. However, when I try and remove the second proxy pass (for /), the page no longer works. Any idea whythis might be? (I can send additional error log info).
How are you accessing your server? If the proxy pass is for /, then you should be able to access your server via yourserver.org/, but if the proxy pass is under /catmaid/, then try typing yourserver.org/catmaid/ in your browser.
Best regards,
Sanja
Marcel Sayre
Sep 2, 2019, 11:11:58 AM9/2/19
to cat...@googlegroups.com
Hey Sanja and Tom,
Sanja, I typically access my server using localhost/catmaid/, but I've just tried localhost/ and that works as well. So perhaps, I can get rid of that block (with the / proxy pass) and just use localhost/catmaid/ to access the server?
Also Tom, regarding my most recent question in red above, I've figured out the issue regarding transferring my .SWC skeletons from Amira into CATMAID. My original Amira stack was slightly offset in the y direction, so the only thing I needed to apply was that offset in the CATMAID translation field.
Thanks for both of your help! I'm sure I'll have another post here soon. ;)
warm regards,
Marcel
--
You received this message because you are subscribed to the Google Groups "CATMAID" group.
To unsubscribe from this group and stop receiving emails from it, send an email to catmaid+u...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/catmaid/73053d87-f875-42fc-abd4-b26f8c25aca8%40googlegroups.com.
Reply all
Reply to author
Forward
0 new messages
