New development release available (3.99_002)
41 views
Skip to first unread message
Shawn Laffan
Nov 17, 2021, 1:06:38 AM11/17/21
to biodiver...@googlegroups.com
A new development release of Biodiverse is now available. This is
the second in the 3.99 series, leading to a version 4 release.
You can access it through https://github.com/shawnlaffan/biodiverse/wiki/Downloads#development-release
An executable is available only for Windows at this time. Mac and Linux releases can be made if needed.
More details and links relating to the new developments can be accessed at https://github.com/shawnlaffan/biodiverse/wiki/ReleaseNotes#version-399-dev-series
Please give it a test run and report any errors or suggest any improvements. You can use this email list or the project issue tracker at https://github.com/shawnlaffan/biodiverse/issues
The main highlights since 3.99_001 are:
New conditions
Cluster and RegionGrower analyses can be exported to shapefile format in a grouped form. This models the cluster display where multiple sub-clusters are coloured. The output file needs further processing outside Biodiverse to be easily used, which is why the sp_points_in_same_cluster() spatial condition was developed (see previous point).
Regards,
Shawn.
You can access it through https://github.com/shawnlaffan/biodiverse/wiki/Downloads#development-release
An executable is available only for Windows at this time. Mac and Linux releases can be made if needed.
More details and links relating to the new developments can be accessed at https://github.com/shawnlaffan/biodiverse/wiki/ReleaseNotes#version-399-dev-series
Please give it a test run and report any errors or suggest any improvements. You can use this email list or the project issue tracker at https://github.com/shawnlaffan/biodiverse/issues
The main highlights since 3.99_001 are:
New conditions
sp_point_in_cluster() and sp_points_in_same_cluster()
that can be used to model polygons from clusters in a Cluster or
RegionGrower analysis. This avoids the need to first export them to
a shapefile and then process that file to extract and dissolve the
relevant parts. Cluster and RegionGrower analyses can be exported to shapefile format in a grouped form. This models the cluster display where multiple sub-clusters are coloured. The output file needs further processing outside Biodiverse to be easily used, which is why the sp_points_in_same_cluster() spatial condition was developed (see previous point).
Regards,
Shawn.
wyckliffe
Jun 11, 2022, 2:56:25 PM6/11/22
to biodiver...@googlegroups.com
Hello , I have a question
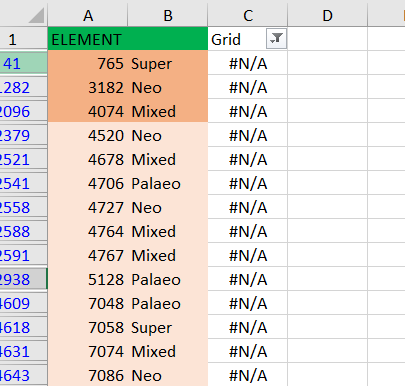
Lately I have been doing CANAPE for a large data set in biodiverse 3.1. Am missing some grids in my original data, for example, grid 700 but I have grid 7001, but after running the spatial analysis and the randomization, I realize that my result is having the missing grid cells.
What could be the possible explanation for this and can it affect the credibility of my CANAPE results.
Attached is a screenshot of my concern. N/A means that these grid cells were missing in my original data (distribution data), but after doing all the CANAPE steps carefully , I noticed these grid cells have been assigned different types of endemism.
Kindly help me
Thank you
Wyckliffe

--
You received this message because you are subscribed to the Google Groups "Biodiverse Users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to biodiverse-use...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/biodiverse-users/86fc2d05-24d1-4bd5-6e94-cbc3446c79e7%40unsw.edu.au.
Shawn Laffan
Jun 11, 2022, 7:05:50 PM6/11/22
to biodiver...@googlegroups.com, wyckliffe
Hello Wyckliffe,
If a group (cell) does not exist in your basedata then Biodiverse will not assign any values to it.
Do these groups exist in Biodiverse when you plot the data?
The issue might be in the step where the CANAPE classes are assigned, as this is done outside Biodiverse. If you are using excel then things can become misaligned. Can you provide more details about how you are doing this?
Regards,
Shawn.
If a group (cell) does not exist in your basedata then Biodiverse will not assign any values to it.
Do these groups exist in Biodiverse when you plot the data?
The issue might be in the step where the CANAPE classes are assigned, as this is done outside Biodiverse. If you are using excel then things can become misaligned. Can you provide more details about how you are doing this?
Regards,
Shawn.
To view this discussion on the web visit https://groups.google.com/d/msgid/biodiverse-users/CAFq2Z%2B19e7ECSNXev_MRrhbjr-SRWTkqzkm0O4%3Dpj3WFve5waQ%40mail.gmail.com.
-- Prof Shawn Laffan, FMSSANZ Director, Earth and Sustainability Science Research Centre School of Biological, Earth and Environmental Sciences UNSW, Sydney 2052, Australia Tel +61 2 9065 5607 https://www.bees.unsw.edu.au/our-people/shawn-laffan https://www.unsw.edu.au/research/essrc https://shawnlaffan.github.io/biodiverse (free diversity analysis software) International Journal of Geographical Information Science http://www.tandf.co.uk/journals/ijgis
Shawn Laffan
Jun 13, 2022, 2:04:32 AM6/13/22
to biodiver...@googlegroups.com
Just an update for the list that this has been resolved.
The data did contain records associated with these grid cells. The issue was that they were beyond the display limits of Excel when the input CSV file was checked so were not visible. Only the first 1,048,576 records were shown.
Spreadsheet programs are very useful to see CSV files with the data formatted in columns, but text editors are very handy when one just needs to search for text strings. There are also unix tools like grep if one is comfortable working on a command line.
Another approach to check if groups exist is to use Biodiverse to transpose the basedata. This is the same idea as transposing a site by species matrix. When the transposed basedata is opened in a View Labels tab it will list all the group names from the original basedata. These can then be searched and selected like any other basedata - https://biodiverse-analysis-software.blogspot.com/2014/12/label-selections-in-view-labels-tab.html
Shawn.
The data did contain records associated with these grid cells. The issue was that they were beyond the display limits of Excel when the input CSV file was checked so were not visible. Only the first 1,048,576 records were shown.
Spreadsheet programs are very useful to see CSV files with the data formatted in columns, but text editors are very handy when one just needs to search for text strings. There are also unix tools like grep if one is comfortable working on a command line.
Another approach to check if groups exist is to use Biodiverse to transpose the basedata. This is the same idea as transposing a site by species matrix. When the transposed basedata is opened in a View Labels tab it will list all the group names from the original basedata. These can then be searched and selected like any other basedata - https://biodiverse-analysis-software.blogspot.com/2014/12/label-selections-in-view-labels-tab.html
Shawn.
To view this discussion on the web visit https://groups.google.com/d/msgid/biodiverse-users/546d2c9c-d85e-89f8-14b4-9facfb288e72%40unsw.edu.au.
Reply all
Reply to author
Forward
0 new messages
