Assembly file contains multiple entries of the same contig. Exiting!
106 views
Skip to first unread message
Zhaoxi Liu
Jul 30, 2021, 4:16:25 AM7/30/21
to 3D Genomics
Hello everyone,
When I run <run-asm-pipeline.sh> using this command "run-asm-pipeline.sh -r 5 -m diploid --editor-coarse-norm "NONE" ../references/mel_contigs_pilon.fasta ../aligned/merged_nodups.txt "
I got this errer " :( Assembly file contains multiple entries of the same contig. Exiting! "
and also " tail: +: invalid number of bytes
tail: +: invalid number of bytes
tail: +: invalid number of bytes
…… "
the vesion is 3d-dna/190716
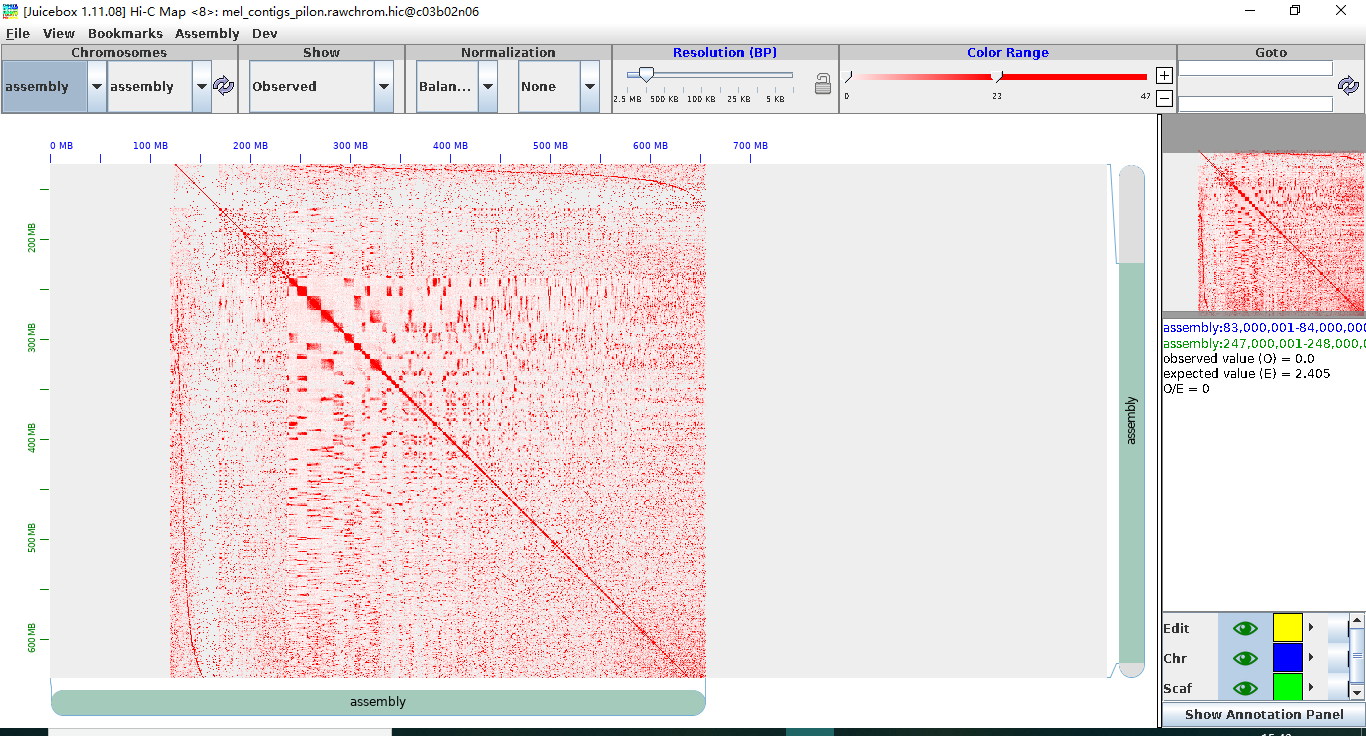
Although it seems that I got all the files including rawchrom.hic and final .assembly and so on, when I use JBAT to open them, there are not large scaffolds but small fragments. The scaffolds are even smaller than the contigs before the assembly. Could you tell me what the problems are? thanks a lot !!!
Best wishes,
Zhaoxi
Zhaoxi Liu
Jul 30, 2021, 4:18:56 AM7/30/21
to 3D Genomics
Olga Dudchenko
Aug 26, 2021, 4:01:34 PM8/26/21
to 3D Genomics
Hi Liu,
Please note that we no longer offer support to the diploid workflow. You'd have to use legacy versions if you want to run in diploid mode, and I would not certainly assume it from the getgo.
Before moving on to reviewing rawchrom files our recommendation is always to examine .0.hic files and tracks.
Apologies for the delayed response.
Thanks,
Olga
On Friday, July 30, 2021 at 3:18:56 AM UTC-5 liuzha...@gmail.com wrote:
Zhaoxi Liu
Aug 27, 2021, 8:50:12 AM8/27/21
to 3D Genomics
Hi
Dudchenko,
Thank you for your advice!
We realized the serious undercollapsed heterozygocity and changed the assembly software HiCanu to wtdbg2 to reassemble the contigs. And then we used the haploid mode of 3D-DNA to scaffold. Now the problems have been solved.
Thanks again and best wishes.
Zhaoxi
Reply all
Reply to author
Forward
0 new messages
