Re: Question on the bifactor invariance modeling
Phil Chalmers
Hi Phil,
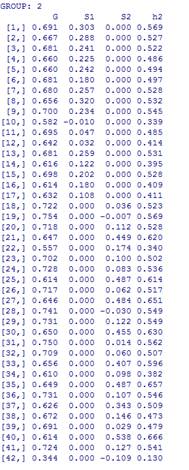
I noticed that when I tested strong factorial invariance (equal slopes and intercepts) using bfactor function, the results indicated only the group factor loadings were set equal between 2 groups. General factor loadings were different. Is this normal? Please see the screenshots below. I didn’t print out all the items. I thought that the highlighted part should have taken care of the issue, but apparently it did not. The code I used is as follows.
group_factor <- c(1,1,1,1,1,1,1,1,1,1,
1,1,1,1,1,1,1,2,2,2,
2,2,2,2,2,2,2,2,2,2,
2,2,2,2,2,2,2,2,2,2,
2,2,2,2,2,2,1,1,2,2,
1,2,2,2,2,2,2,1,1,1,
1,1,1,1,1,1,1,1,1,1,
1,1,1,2,2,2,2,2,2,2,
2,2,2,2,2,2,2,2,2,2,
2,1,1,1,1)
inv.model2 <- bfactor(combined.data,
model=group_factor,
invariance= c("slopes", "intercepts","free_var","free_means"),
itemtype="graded",
group=grp,
quadpts = 32,
verbose=TRUE,
TOL=1E-05,
technical=list(NCYCLES=5000))
Thank you for your consideration!
Minji
Group 1
Here is group 2