Cannot categorize reads by "sample"
66 views
Skip to first unread message
Balthazar Picsou
Mar 17, 2017, 6:44:37 PM3/17/17
to igv-help, rwi...@pha.jhu.edu
Hello --
I cannot get IGV to categorize reads by "sample" from a SAM file. The IGV docs say:
"Sample is a tag designated in the SAM format file header section under @RG that specifies sample information. E.g., a sample may by split and run across multiple sequencing lanes represented by different read groups but the same sample tag."
Ok, fine, but when I display the reads in an IGV track, right click on the track, and select "Group alignments by... sample" (or "Color" or "Sort"), nothing happens.
I have attached a little SAM file to demonstrate this. What is wrong here? Is it the SAM file contents? (If so, I don't see the problem.) Or is it something I am doing (or not doing) with IGV?
Thanks!
I cannot get IGV to categorize reads by "sample" from a SAM file. The IGV docs say:
"Sample is a tag designated in the SAM format file header section under @RG that specifies sample information. E.g., a sample may by split and run across multiple sequencing lanes represented by different read groups but the same sample tag."
Ok, fine, but when I display the reads in an IGV track, right click on the track, and select "Group alignments by... sample" (or "Color" or "Sort"), nothing happens.
I have attached a little SAM file to demonstrate this. What is wrong here? Is it the SAM file contents? (If so, I don't see the problem.) Or is it something I am doing (or not doing) with IGV?
Thanks!
James Robinson
Mar 17, 2017, 8:40:14 PM3/17/17
to igv-help
What version of IGV are you using?
--
---
You received this message because you are subscribed to the Google Groups "igv-help" group.
To unsubscribe from this group and stop receiving emails from it, send an email to igv-help+unsubscribe@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/igv-help/46122795-28b9-433d-bf99-4c5f2176ce7a%40googlegroups.com.
For more options, visit https://groups.google.com/d/optout.
Balthazar Picsou
Mar 27, 2017, 7:06:01 PM3/27/17
to igv-help
2.3.69
On Friday, March 17, 2017 at 5:40:14 PM UTC-7, Jim Robinson wrote:
What version of IGV are you using?
On Fri, Mar 17, 2017 at 3:44 PM, 'Balthazar Picsou' via igv-help <igv-...@googlegroups.com> wrote:
Hello --
I cannot get IGV to categorize reads by "sample" from a SAM file. The IGV docs say:
"Sample is a tag designated in the SAM format file header section under @RG that specifies sample information. E.g., a sample may by split and run across multiple sequencing lanes represented by different read groups but the same sample tag."
Ok, fine, but when I display the reads in an IGV track, right click on the track, and select "Group alignments by... sample" (or "Color" or "Sort"), nothing happens.
I have attached a little SAM file to demonstrate this. What is wrong here? Is it the SAM file contents? (If so, I don't see the problem.) Or is it something I am doing (or not doing) with IGV?
Thanks!
--
---
You received this message because you are subscribed to the Google Groups "igv-help" group.
To unsubscribe from this group and stop receiving emails from it, send an email to igv-help+u...@googlegroups.com.
James Robinson
Mar 27, 2017, 10:14:19 PM3/27/17
to igv-help
Please update to the latest version and try again. Also, if reporting a bug against an old version of IGV it's helpful if you note that.
Balthazar Picsou
Mar 29, 2017, 12:09:34 PM3/29/17
to igv-help, rwi...@pha.jhu.edu
Ok, fine, I downloaded v2.3.92 (146) and got the same goofy results. Perhaps the advice to "update to the latest version and try again" was simply generic advice and not based on any known bug fix.
Anyway.
For what it's worth, sorting/grouping/coloring the mappings by read group seems to work ok with the test data I uploaded.
James Robinson
Mar 29, 2017, 2:24:57 PM3/29/17
to igv-help, rwi...@pha.jhu.edu
It wasn't generic advice, I tried your file with 2.3.92 and it worked fine, I was able to group and color by sample. See screenshot
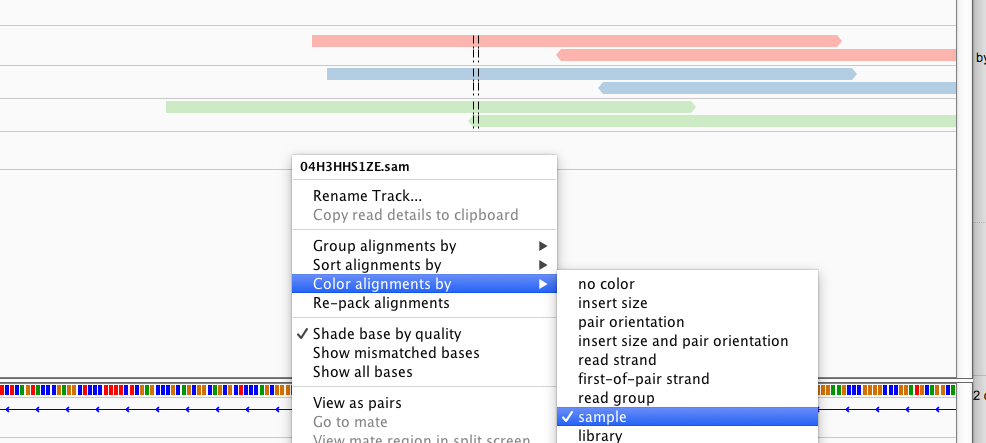

--
---
You received this message because you are subscribed to the Google Groups "igv-help" group.
To unsubscribe from this group and stop receiving emails from it, send an email to igv-help+unsubscribe@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/igv-help/34d5d19d-828e-4060-965f-7da0b5c3139d%40googlegroups.com.
Balthazar Picsou
Mar 29, 2017, 5:54:19 PM3/29/17
to igv-help, rwi...@pha.jhu.edu
That ought to help narrow down the problem. See screenshot.
I have also uploaded a copy of the startup log, if that helps at all.
On Wednesday, March 29, 2017 at 11:24:57 AM UTC-7, Jim Robinson wrote:
It wasn't generic advice, I tried your file with 2.3.92 and it worked fine, I was able to group and color by sample. See screenshot
On Wed, Mar 29, 2017 at 9:09 AM, 'Balthazar Picsou' via igv-help <igv-...@googlegroups.com> wrote:
Ok, fine, I downloaded v2.3.92 (146) and got the same goofy results. Perhaps the advice to "update to the latest version and try again" was simply generic advice and not based on any known bug fix.
Anyway.
For what it's worth, sorting/grouping/coloring the mappings by read group seems to work ok with the test data I uploaded.
On Friday, March 17, 2017 at 3:44:37 PM UTC-7, Balthazar Picsou wrote:Hello --
I cannot get IGV to categorize reads by "sample" from a SAM file. The IGV docs say:
"Sample is a tag designated in the SAM format file header section under @RG that specifies sample information. E.g., a sample may by split and run across multiple sequencing lanes represented by different read groups but the same sample tag."
Ok, fine, but when I display the reads in an IGV track, right click on the track, and select "Group alignments by... sample" (or "Color" or "Sort"), nothing happens.
I have attached a little SAM file to demonstrate this. What is wrong here? Is it the SAM file contents? (If so, I don't see the problem.) Or is it something I am doing (or not doing) with IGV?
Thanks!
--
---
You received this message because you are subscribed to the Google Groups "igv-help" group.
To unsubscribe from this group and stop receiving emails from it, send an email to igv-help+u...@googlegroups.com.
James Robinson
Mar 29, 2017, 9:55:37 PM3/29/17
to igv-help
Thank you for the screenshot, it was helpful. There is indeed a problem and I can reproduce it now. Will update this thread when I find the cause.
To unsubscribe from this group and stop receiving emails from it, send an email to igv-help+unsubscribe@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/igv-help/1e7ecb11-9679-430c-99dc-d8d4b66a0c9c%40googlegroups.com.
James Robinson
Mar 30, 2017, 11:45:09 AM3/30/17
to igv-help
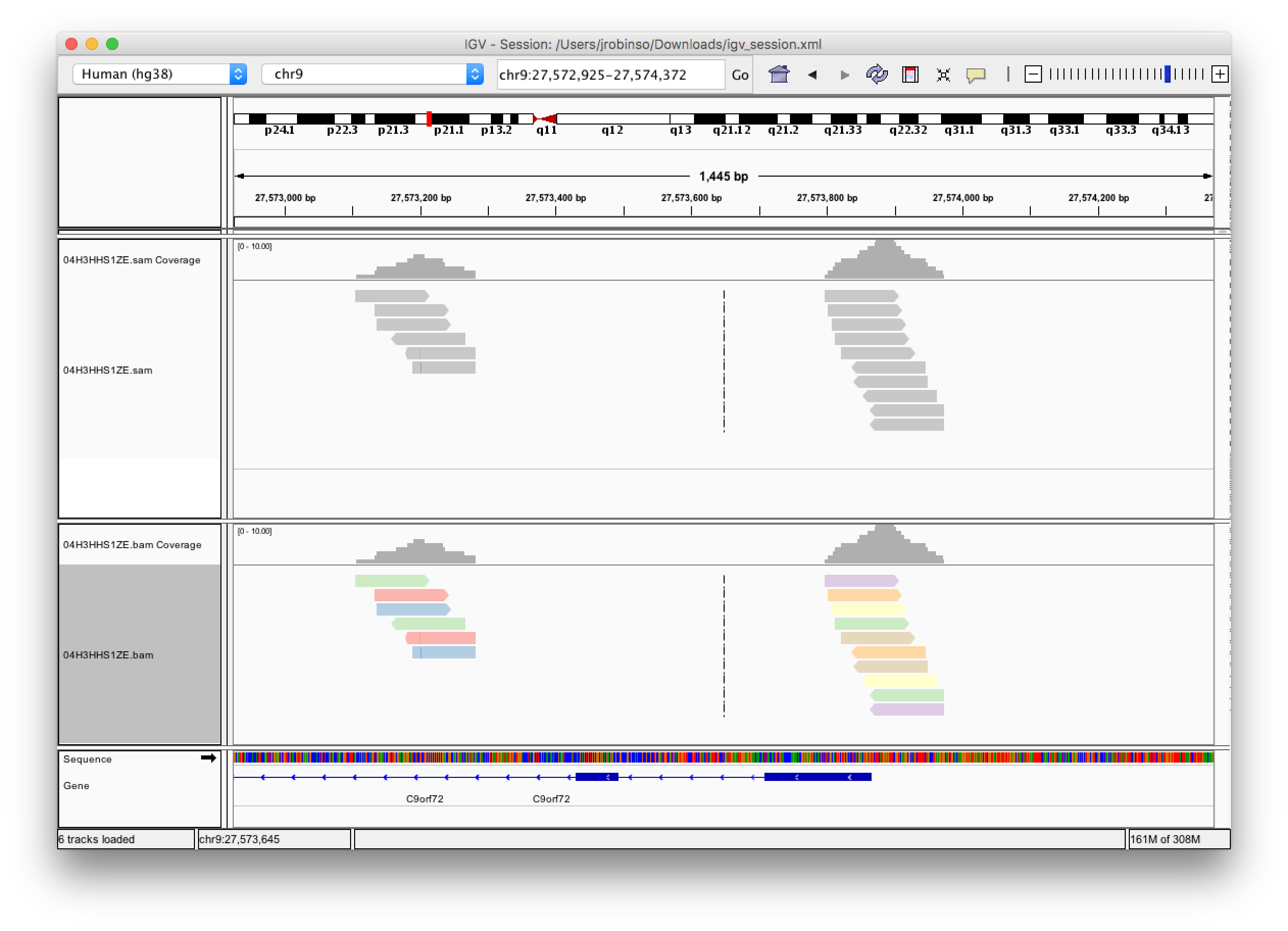
OK, I found the cause of this. It was difficult to track down, but there is a bug in the htsjdk library. When reading plain text "SAM" files the header is not completely or correctly parsed at some resolutions. There isn't anything we can do about this in IGV. The fix is to use ".bam" files. Screenshot is your example file in sam and bam format, both panels have "color by sample" on.

Balthazar Picsou
Mar 31, 2017, 12:31:14 AM3/31/17
to igv-help
Good to know that you found the bug, and thank you for proposing a workaround. It's frustrating that it's not readily fixable. Do I need to pass this on to the samtools folks?
By the way, I am puzzled by the dependence of a SAM parsing error on the granularity of the visual representation of the reference genome. That's not an "intuitive" relationship, at least not from the outside where I sit!
James Robinson
Mar 31, 2017, 1:20:02 AM3/31/17
to igv-help
You can file this as an htsjdk bug, but I know parsing of SAM files is not a priority for them. They recommend BAM for everything. The bug is strange, if you start out very zoomed in (40 bp window) and load the sam records the "read group" records are present, that's why it worked for me originally. However, if you start with the locus shown in the figure, a 1500 bp window, the read group records are not present in the "SAMRecord" object from the htsjdk. I stepped through this in the htsjdk and verified that they really aren't there. The "RG" tag is present, that's why that still works, but the read group object is not, so sample is "null". Anyway that's as far as I got, IGV's support for SAM text file parsing is legacy and old, even the index is not an htsjdk supported feature, there just isn't much support anywhere for text SAM files.
To unsubscribe from this group and stop receiving emails from it, send an email to igv-help+unsubscribe@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/igv-help/52961eda-5907-495d-84f8-2d6a99dcd063%40googlegroups.com.
Message has been deleted
Kirk McClure
Jul 4, 2017, 8:35:57 PM7/4/17
to igv-help
The header field of objects returned by the htsjdk SAMTextReader.RecordIterator.next() method is not fully populated.
An admittedly blunt work-around in the IGV SAMQueryIterator class corrects the issue when tested with the referenced .sam file.
import htsjdk.samtools.SAMFileHeader;
...
SAMFileHeader mHeader; // (global)
...
in SAMQueryIterator() // (two variants)
mHeader = null;
...
conditionally replace the header in next()
public PicardAlignment next() {
SAMRecord ret = currentRecord;
if (wrappedIterator.hasNext()) {
currentRecord = wrappedIterator.next();
if (mHeader != null) currentRecord.setHeader( mHeader ); // addition
} else {
currentRecord = null;
}
return new PicardAlignment(ret);
}
...
new method:
public void setHeader( SAMFileHeader header ) {
mHeader = header.clone();
if ( currentRecord != null ) currentRecord.setHeader( mHeader );
}
and in IGV SAMReader query()
replace
return new SAMQueryIterator(sequence, start, end, contained, iter);
with
SAMQueryIterator sqi = new SAMQueryIterator(sequence, start, end, contained, iter);
sqi.setHeader( header );
return sqi;
Reply all
Reply to author
Forward
0 new messages