adding information on default reftable annotation
22 views
Skip to first unread message
Varun Gupta
Jun 26, 2017, 5:28:40 PM6/26/17
to igv-help
Hi,
I am trying to make sashimi plots for my gene of interest. This gene has a micro-exon which is not annotated in hg19 ref seq reftable which igv uses by default. How can I add that micro-exon information in the default annotation which IGV uses? This is because, if IGV doesn't know about this exon, it won't show any splicing on the shashimi plot.
Hope to hear from you soon.
Regards
Varun
I am trying to make sashimi plots for my gene of interest. This gene has a micro-exon which is not annotated in hg19 ref seq reftable which igv uses by default. How can I add that micro-exon information in the default annotation which IGV uses? This is because, if IGV doesn't know about this exon, it won't show any splicing on the shashimi plot.
Hope to hear from you soon.
Regards
Varun
James Robinson
Jun 26, 2017, 5:59:11 PM6/26/17
to igv-help
You can load your own annotation file as bed, gtf, or gff. When opening the Sashimi plot it lets you choose which annotation track to use.
--
---
You received this message because you are subscribed to the Google Groups "igv-help" group.
To unsubscribe from this group and stop receiving emails from it, send an email to igv-help+unsubscribe@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/igv-help/613546fb-d9e6-4482-9ae9-824f9f9398d7%40googlegroups.com.
For more options, visit https://groups.google.com/d/optout.
Varun Gupta
Jun 27, 2017, 12:03:50 PM6/27/17
to igv-help
Thanks JIm. That worked
Regards
Varun
Regards
Varun
On Monday, June 26, 2017 at 5:59:11 PM UTC-4, Jim Robinson wrote:
You can load your own annotation file as bed, gtf, or gff. When opening the Sashimi plot it lets you choose which annotation track to use.
On Mon, Jun 26, 2017 at 2:28 PM, Varun Gupta <gupta5...@gmail.com> wrote:
Hi,
I am trying to make sashimi plots for my gene of interest. This gene has a micro-exon which is not annotated in hg19 ref seq reftable which igv uses by default. How can I add that micro-exon information in the default annotation which IGV uses? This is because, if IGV doesn't know about this exon, it won't show any splicing on the shashimi plot.
Hope to hear from you soon.
Regards
Varun
--
---
You received this message because you are subscribed to the Google Groups "igv-help" group.
To unsubscribe from this group and stop receiving emails from it, send an email to igv-help+u...@googlegroups.com.
Varun Gupta
Jun 27, 2017, 3:07:13 PM6/27/17
to igv-help
Hi Jim,
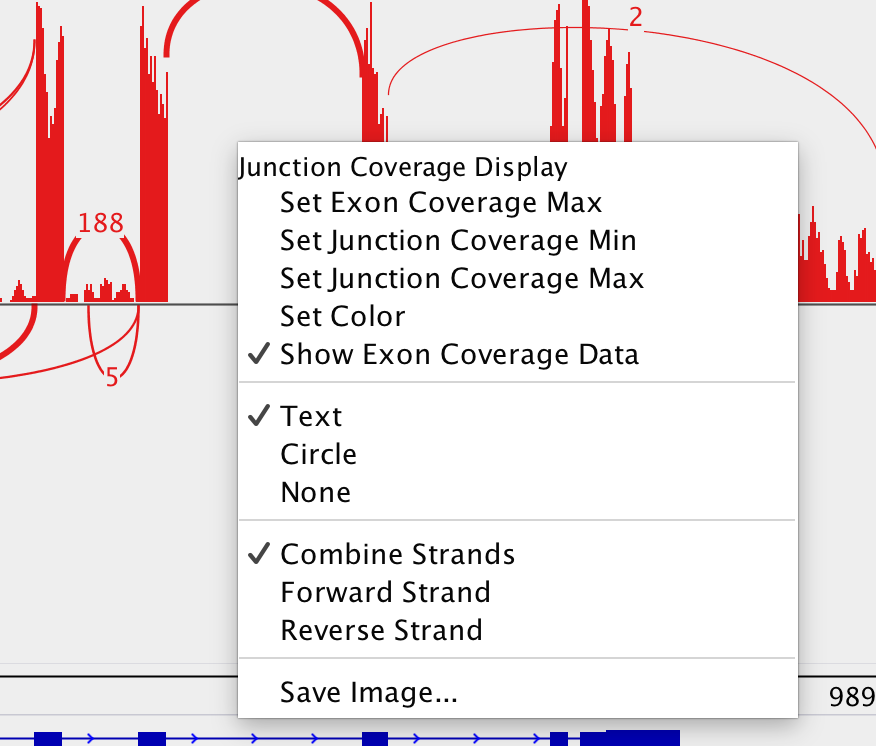
I was able to modify the refFlat file and was able to load it with my micro-exon. When doing Sashimi plot though, I cannot see the junction coverage across that micro-exon, even though we have lot of junctions around it.
What options should I change.
Thanks for the help.
Regards
Varun
On Monday, June 26, 2017 at 5:59:11 PM UTC-4, Jim Robinson wrote:
On Monday, June 26, 2017 at 5:59:11 PM UTC-4, Jim Robinson wrote:
You can load your own annotation file as bed, gtf, or gff. When opening the Sashimi plot it lets you choose which annotation track to use.
On Mon, Jun 26, 2017 at 2:28 PM, Varun Gupta <gupta5...@gmail.com> wrote:
Hi,
I am trying to make sashimi plots for my gene of interest. This gene has a micro-exon which is not annotated in hg19 ref seq reftable which igv uses by default. How can I add that micro-exon information in the default annotation which IGV uses? This is because, if IGV doesn't know about this exon, it won't show any splicing on the shashimi plot.
Hope to hear from you soon.
Regards
Varun
--
---
You received this message because you are subscribed to the Google Groups "igv-help" group.
To unsubscribe from this group and stop receiving emails from it, send an email to igv-help+u...@googlegroups.com.
James Robinson
Jul 10, 2017, 12:05:19 PM7/10/17
to igv-help
I assume you have "Show Exon Coverage Data" checked? I don't understand your comment about "junctions around it", the only thing that matters are reads that align or partially align to that exon. If you don't see look at the coverage of the exon on the regular alignment track, is there any there? If so check the data range on the sashimi plot, perhaps the max value is too large. You can set it by right-clicking and selecting "Set Exon Coverage Max".

On Tue, Jun 27, 2017 at 12:07 PM, Varun Gupta <gupta5...@gmail.com> wrote:
Hi Jim,I was able to modify the refFlat file and was able to load it with my micro-exon. When doing Sashimi plot though, I cannot see the junction coverage across that micro-exon, even though we have lot of junctions around it.What options should I change.
Reply all
Reply to author
Forward
0 new messages
