Adding second legend to ggplot
Kristina Wolf
Matt Whalen
Matt Whalen
PhD Candidate, Graduate Group in Ecology
Department of Evolution & Ecology
University of California, Davis
Bodega Marine Laboratory
--
Check out our R resources at http://d-rug.github.io/
---
You received this message because you are subscribed to the Google Groups "Davis R Users' Group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to davis-rug+...@googlegroups.com.
Visit this group at https://groups.google.com/group/davis-rug.
For more options, visit https://groups.google.com/d/optout.
Kristina Michelle Wolf
Thanks so much Matt!
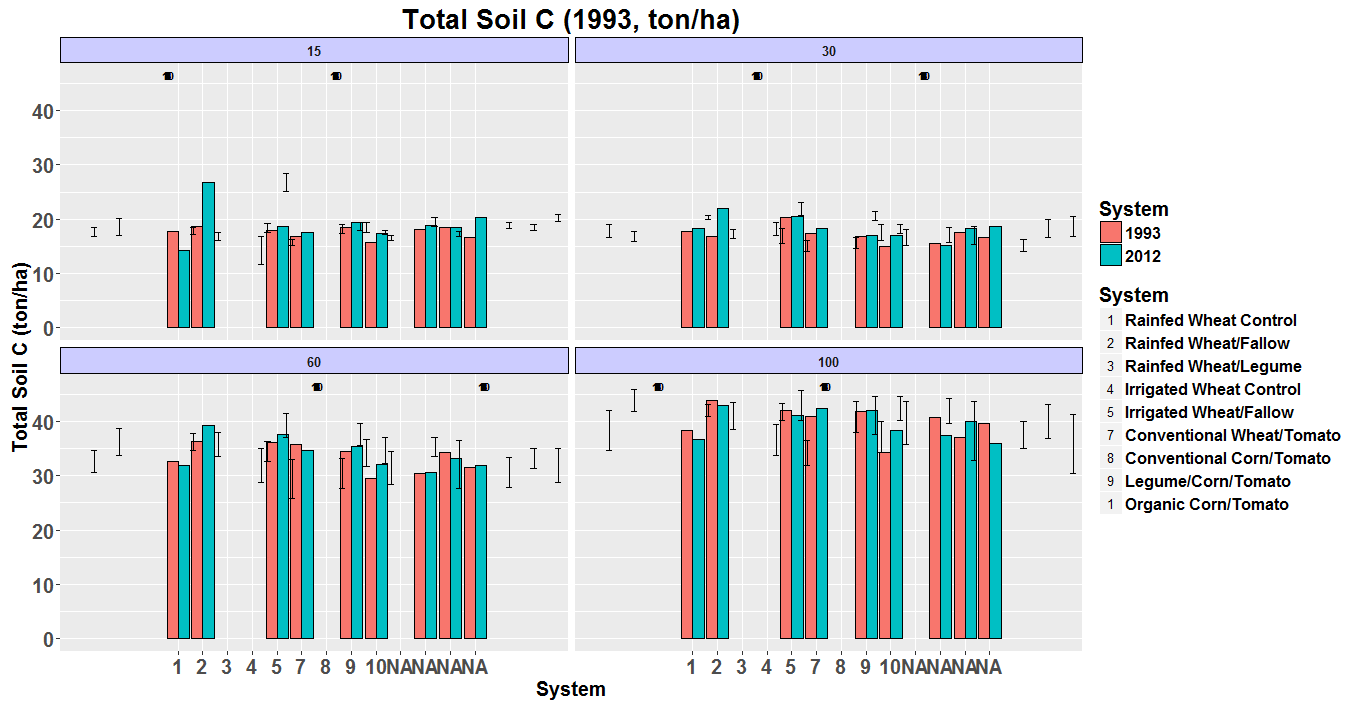
Getting closer, but it’s pretty wonky at this point still. I am going to include the entire code, including some commented out portions so you can see things. The system number and year are already made into factors, so that should be ok to start, but…
Notice that the System legend goes from 1:5, 7:9, and back to 1. It is fine that it skips 6, because there is no System 6, but it is dropping the zero on System 10. I tried editing the code you sent to specifically call them (see the code, rather than “labels = 1:10”, I used “labels = c(all the system numbers in quotes)” to see if that would help, and no-go,
I also have a bunch of NAs on the x-axis, and the labels and error bars are all over the place:
ggplot(CN_by_year, aes(x=Treatment.Number, y = mean_c, fill = year)) +
geom_bar(position="dodge", stat = "identity", colour = "black") +
geom_errorbar(CN_by_year, mapping = aes(ymin=mean_c-se_c, ymax=mean_c+se_c), width=0.5, position=position_dodge(.9*15)) +
facet_wrap(~Lower.Depth, nrow=2) +
# scale_x_discrete(name="System", breaks=c("1", "2", "3", "4", "5", "7", "8", "9", "10"), labels = c("Rainfed Wheat Control", "Rainfed Wheat/Fallow", "Rainfed Wheat/Legume", "Irrigated Wheat Control", "Irrigated Wheat/Fallow", "Conventional Wheat/Tomato", "Conventional Corn/Tomato", "Legume/Tomato", "Organic Corn/Tomato")) +
scale_x_discrete(name="System", labels=c("1","2","3","4","5","7","8","9","10")) +
geom_point(aes(shape = Treatment.Number), alpha = 0) +
guides(shape = guide_legend(override.aes = list(size = 3, alpha = 1))) +
scale_shape_manual(name = "System",values=levels(CN_by_year$Treatment.Number),labels=levels(CN_by_year$Treatment.Name)) +
scale_fill_manual(values=c("#CCCCCC", "#333333"),
name="Year",
breaks = c("1993", "2012"),
labels=c("1993", "2012")) +
xlab("System") +
scale_y_continuous(name = "Total Soil C (ton/ha)") +
ggtitle("Total Soil C (1993, ton/ha)") +
scale_fill_hue(name="System") +
geom_text(CN_by_year, mapping = aes(x=Lower.Depth , y = max(mean_c+se_c+0.55, mean_c+0.55),label=Treatment.Number), size = 3, position=position_dodge(.9*15), fontface = "bold") +
theme(plot.title = element_text(face="bold", size=20),
axis.title.x = element_text(face="bold", size=15),
axis.text.x = element_text(face="bold", size=15),
axis.title.y = element_text(face="bold", size=15),
axis.text =element_text(face="bold", size=15),
strip.text.x = element_text(face = "bold", size=10),
strip.background = element_rect(colour="black", fill="#CCCCFF"),
legend.title = element_text(face ="bold", size=15),
legend.text = element_text(face = "bold", size = 12))
Kristina Wolf
Research & Academic Coordinator
Russell Ranch
Agricultural Sustainability Institute
U.C. Davis
Ph.D. Ecology, M.S. Soil Science, B.S. Animal Science
Laboratory (530) 754-5203
Cellular (530) 574-9102

Ryan Peek
John Muir (My First Summer in the Sierra, 1911)
