Filtering a Factor Field in dplyr
20 views
Skip to first unread message
Nick Santos
Mar 22, 2018, 12:24:11 AM3/22/18
to davi...@googlegroups.com
Hi all,
I have a kind of weird situation. I have a presence absence matrix looking something like:"180105010201", 0, 1, ..., 0, 1
"180105010202", 1, 1, ..., 0, 3
huc_data <- st_as_sf(readOGR("path_to_geopackage", "layer_name"))
group = 1 # for example
records_in_group <- huc_data %>% filter(group_id == group)
# or while debugging, to be explicit
records_in_group <- as.data.frame(huc_data) %>% dplyr::filter(group_id == as.character(group))
# I've also tried just putting group_id == 1 and group_id == "1" for testing
Whenever I run the filtering
code, I get an empty data frame, though I can verify that it has records that match the query as I read it. I suspect the issue is that the fields are factors and I'm somehow failing to account for that in the code I'm writing, but I'm not positive. It could also be related to it coming in as an SF geometry first, but by the time I'm filtering it, everything else about it having been SF should be gone given the call I mentioned earlier. See below for a sample part of the schema.
The top 3 are species fields and the bottom two would be group_id fields.
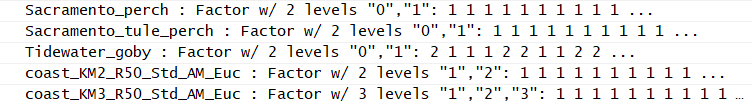

This is all running in R 3.4.0 on Windows 10, with dplyr 0.7.4 and sf 0.6-0 (coming from GitHub, not from CRAN)
Katherine Ransom
Mar 22, 2018, 11:39:48 AM3/22/18
to davis-rug
Hi Nick,
Is huc_ data a data.frame then? What do you get when you call class() on it?
Best,
Katie
--
Check out our R resources at http://d-rug.github.io/
---
You received this message because you are subscribed to the Google Groups "Davis R Users' Group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to davis-rug+...@googlegroups.com.
Visit this group at https://groups.google.com/group/davis-rug.
For more options, visit https://groups.google.com/d/optout.
Nick Santos
Mar 22, 2018, 12:02:27 PM3/22/18
to davi...@googlegroups.com
Hi Katherine,
Thanks for the reply - yes, it is.
At first it's dual-classed as an sf object and a data frame, but by the
time I'm doing the filtering, I've dropped the SF geometry and it shows
as just a data.frame:
> class(huc_data) [1] "sf" "data.frame"
then later, I get
> class(data_of_interest) # this is the variable that's actually being passed into the filter call, but I kept it simpler in my question [1] "data.frame"
That said, maybe there's a
better way for me to drop the geometry or something? Reading the docs,
it looks like as.data.frame should do it just fine too, but testing that
while skipping the call where I explicitly set the geometry to null
doesn't result in anything. Looking at the docs, I also see that there's
a filter method for SF objects, but that's not working either (Error in NextMethod() : generic function not specified - chasing this down, I only get recommendations to do things I'm already doing).
Thanks again!
To unsubscribe from this group and stop receiving emails from it, send an email to davis-rug+unsubscribe@googlegroups.com.
Visit this group at https://groups.google.com/group/davis-rug.
For more options, visit https://groups.google.com/d/optout.
--
Check out our R resources at http://d-rug.github.io/
---
You received this message because you are subscribed to the Google Groups "Davis R Users' Group" group.
To unsubscribe from this group and stop receiving emails from it, send an email to davis-rug+unsubscribe@googlegroups.com.
Scott Devine
Mar 22, 2018, 12:37:31 PM3/22/18
to davi...@googlegroups.com
Hi Nick, try changing "as.character(group)" to "as.factor(group)", like this when calling filter:
records_in_group <- as.data.frame(huc_data) %>% dplyr::filter(group_id == as.factor(group))
I don't use dplyr but I think you're intuition is right, and that you are having data class issues. If you don't want these data.frame columns to be read-in as factors, you can set the
stringsAsFactors argument to FALSE within the call to "readOGR". See here:
https://www.rdocumentation.org/packages/rgdal/versions/1.2-16/topics/readOGR
If this doesn't work, please go ahead and share full data and code, and I can try to help you debug.
Scott
Scott Devine
PhD candidate in Soils and Biogeochemistry
Dept of Land, Air, and Water Resources
University of California, Davis
Katherine Ransom
Mar 22, 2018, 12:42:57 PM3/22/18
to davis-rug
Hi Nick,
I agree with Scott. The following example with a numerical factor like yours works fine both ways. Also, always good to call the function from within the packages like Scott did, when things aren’t behaving as expected (e.g. dplyr::filter). That way you are sure you aren’t using a function masked from another package. Best, Katie
library(tidyverse)
glimpse(mtcars)
# make a numerical factor group variable
mtcars$carb <- as.factor(mtcars$carb)
glimpse(mtcars)
class(mtcars)
group1 <- mtcars %>%
filter(carb == 1)
group <- c("1")
group1 <- mtcars %>%
filter(carb == group)
--
Katherine Ransom, PhD
Hydrologic Sciences, UC Davis
Nick Santos
Mar 22, 2018, 1:46:15 PM3/22/18
to davi...@googlegroups.com
Thank you both - it ended up being something else that
I'd left out of the code, thinking it wasn't relevant - I found it in
the process of trying to make a reproducible case to share with you and
testing some other combinations.
I'd been specifying the
field name in the filter command as a string variable because it was
being dynamically produced. If I swap it to# for example
dplyr::filter_(paste(field_name, "==", as.character(group)))
This StackOverflow answer has a few other options too.Also, thanks for the recommendation about being explicit about the dplyr:: - Ryan Peek mentioned something similar to me. I'd prefer to write it that way as well since I'm usually coding in languages where namespaces must be explicit, so I've become accustomed to (and more comfortable) seeing which package a function comes from at a glance.
Katherine Ransom
Mar 22, 2018, 1:58:22 PM3/22/18
to davis-rug
Great! You're welcome! I often find solutions to my R issues the same way, by working up an example to share.
Reply all
Reply to author
Forward
0 new messages
