show chemical structure = error 'transformed width (0) is less than or equal to 0'
28 views
Skip to first unread message
Aidan Schneider
May 14, 2017, 10:26:45 AM5/14/17
to cytoscape-helpdesk
Hi,
I am trying to show chemical structure on my nodes from InChIs but I keep getting the error 'transformed width (0) is less than or equal to 0'
I import a network table that looks like this:
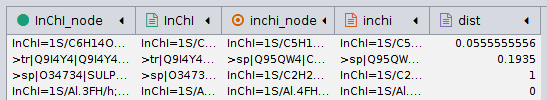
As you can see, it has both inchis (for chemical substrates), sequence headers (for the proteins which act on the substrate), and a distance for the interaction. There are inchi:inchi interactions, sequence:sequence interactions, and inchi:sequence interactions. Since Inchis can be in either column, I labeled one column as InChI and one as inchi to distinguish. The inchi and inchi_node columns are identical.
I select the inchis (as opposed to the sequence headers, like this). I think it also selects the edges connected to inchis (even if they are connected on the other end to sequence headers), but I don't know how to fix that.
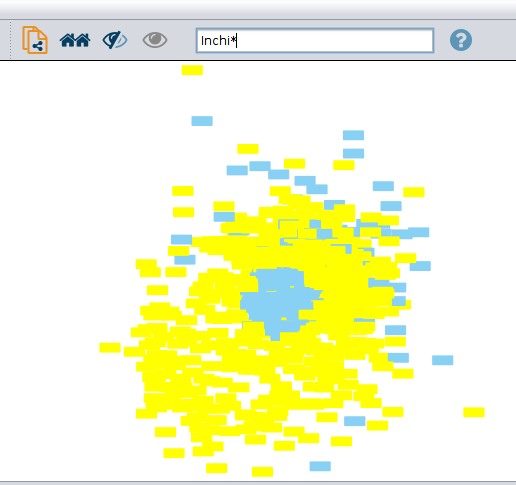
Then, when I click show chemical structure for selected nodes. I get the following error 'transformed width (0) is less than or equal to 0' .
Please help, how can I fix this?
I am trying to show chemical structure on my nodes from InChIs but I keep getting the error 'transformed width (0) is less than or equal to 0'
I import a network table that looks like this:
As you can see, it has both inchis (for chemical substrates), sequence headers (for the proteins which act on the substrate), and a distance for the interaction. There are inchi:inchi interactions, sequence:sequence interactions, and inchi:sequence interactions. Since Inchis can be in either column, I labeled one column as InChI and one as inchi to distinguish. The inchi and inchi_node columns are identical.
I select the inchis (as opposed to the sequence headers, like this). I think it also selects the edges connected to inchis (even if they are connected on the other end to sequence headers), but I don't know how to fix that.
Then, when I click show chemical structure for selected nodes. I get the following error 'transformed width (0) is less than or equal to 0' .
Please help, how can I fix this?
Aidan Schneider
May 14, 2017, 10:29:14 AM5/14/17
to cytoscape-helpdesk
To clarify, I am using ChemViz2 under chemoinformatics tools for this.
Scooter Morris
May 14, 2017, 12:45:56 PM5/14/17
to cytoscape...@googlegroups.com
Hi Aidan,
Can you run this from the terminal and let me know if you get a backtrace? That will help me diagnose this directly. I suspect that ChemViz2 is getting confused by having non-InChi descriptors in the InChi column.
-- scooter
Can you run this from the terminal and let me know if you get a backtrace? That will help me diagnose this directly. I suspect that ChemViz2 is getting confused by having non-InChi descriptors in the InChi column.
-- scooter
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
For more options, visit https://groups.google.com/d/optout.
Aidan Schneider
May 16, 2017, 5:57:12 AM5/16/17
to cytoscape-helpdesk
Hi Dr. Morris,
Thanks for the response. Per your emailed suggestion, I separated the columns like this:

However, when I went to chemoinformatics tools: show structure this time after selecting inchis with search inchi* as previously, all options were grayed out. So I went into settings and did this. No real reason for any of my choices, I just guessed.
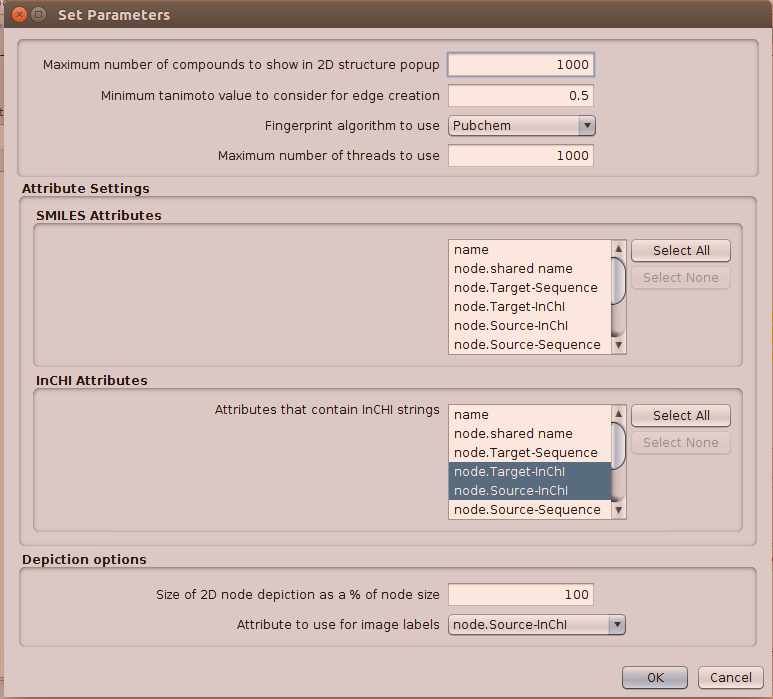
The show structure options appeared now. I chose show structure for selected nodes and still got the same error.
I then searched again with Column name (Source-InChI:*) which selected 247 nodes. Then, I again tried show structure for selected nodes and got the same error.
I ran the program from the terminal with the debug argument and what printed out on the terminal is attached. I've never debugged before, so I'm not sure this is the correct backtrace information you requested. Towards the bottom it has stuff related to the show structure error I think.
Thanks for the help and please let me know what you think.
On Sunday, May 14, 2017 at 5:45:56 PM UTC+1, Scooter Morris wrote:
Thanks for the response. Per your emailed suggestion, I separated the columns like this:
However, when I went to chemoinformatics tools: show structure this time after selecting inchis with search inchi* as previously, all options were grayed out. So I went into settings and did this. No real reason for any of my choices, I just guessed.
The show structure options appeared now. I chose show structure for selected nodes and still got the same error.
I then searched again with Column name (Source-InChI:*) which selected 247 nodes. Then, I again tried show structure for selected nodes and got the same error.
I ran the program from the terminal with the debug argument and what printed out on the terminal is attached. I've never debugged before, so I'm not sure this is the correct backtrace information you requested. Towards the bottom it has stuff related to the show structure error I think.
Thanks for the help and please let me know what you think.
On Sunday, May 14, 2017 at 5:45:56 PM UTC+1, Scooter Morris wrote:
Hi Aidan,
Can you run this from the terminal and let me know if you get a backtrace? That will help me diagnose this directly. I suspect that ChemViz2 is getting confused by having non-InChi descriptors in the InChi column.
-- scooter
On 05/14/2017 07:26 AM, Aidan Schneider wrote:
Hi,
I am trying to show chemical structure on my nodes from InChIs but I keep getting the error 'transformed width (0) is less than or equal to 0'
I import a network table that looks like this:
As you can see, it has both inchis (for chemical substrates), sequence headers (for the proteins which act on the substrate), and a distance for the interaction. There are inchi:inchi interactions, sequence:sequence interactions, and inchi:sequence interactions. Since Inchis can be in either column, I labeled one column as InChI and one as inchi to distinguish. The inchi and inchi_node columns are identical.
I select the inchis (as opposed to the sequence headers, like this). I think it also selects the edges connected to inchis (even if they are connected on the other end to sequence headers), but I don't know how to fix that.
Then, when I click show chemical structure for selected nodes. I get the following error 'transformed width (0) is less than or equal to 0' .
Please help, how can I fix this?
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
Aidan Schneider
May 16, 2017, 5:58:30 AM5/16/17
to cytoscape-helpdesk
Just to clarify, the terminal output is attached as a txt file to the previous post.
On Sunday, May 14, 2017 at 3:26:45 PM UTC+1, Aidan Schneider wrote:
Reply all
Reply to author
Forward
0 new messages
