Generating metabolic network
Ankita Chatterjee
Scooter Morris
In principal, this is relatively easy. All you need to do is to massage your list so that it looks like an edge list:
metabolite1 reaction1 metabolite2
metabolite1 reaction2 metabolite3
The real problem happens when you have more than two metabolites involved in the same reaction. In that case, you need to make your reactions nodes (you can resize the nodes later so they "disappear"). Now your file should look something like:
metabolite1 reaction1 met reac
metabolite2 reaction1 met reac
metabolite1 reaction2 met reac
metabolite3 reaction2 met reac
Now the "met" column should be read in as a source node attribute and the "reac" column should be read in as a target node attribute. This will allow you separately style the metabolites and reaction nodes. By the way, if metabolite3 is a product of reaction2, you would probably write the last like as:
reaction2 metabolite3 reac met
that way you can use edge directions to show the directionality of the reaction.
-- scooter
On 04/27/2017 02:49 AM, Ankita Chatterjee wrote:
I am new to network analysis using Cytoscape. I have names of metabolites and their associated reactions. How to generate a metabolic network in which nodes represent the metabolites and edges represent reactions (So, if two metabolites participate in the same reaction, they should be connected by an edge representing the reaction/enzyme)?
Thank you.
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
For more options, visit https://groups.google.com/d/optout.
Ankita Chatterjee
Scooter Morris
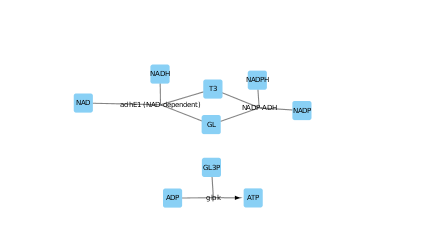
I used the File->Import->Network->File and chose the first column as the source node, the second column as the target, the third column as source node attribute and the final column as a target node attribute. Then I set up a discrete mapper for node size and I set the node size to 1 if the node was a reaction. A little moving things around and I got the above.
-- scooter
Ankita Chatterjee
Thanks again for your time!
On Thursday, April 27, 2017 at 3:19:14 PM UTC+5:30, Ankita Chatterjee wrote:
Scooter Morris
Hi Scooter
I followed the methodology as suggested by you and I could generate a network of the sample file. However, I have few queries: (1) I want the edges to represent reactions. Here, what we are doing is, making the reaction nodes disappear (by assigning node size 1) Thus, edges do not represent the reactions that are common for metabolites.
The reaction names are getting overlapped along the edges and looking clumsy. Should it be the correct representation?
(2) Do we need to assign node size individually to each reaction node manually? In my case the real network will have around 150 metabolites and 400 reactions.
Thanks again for your time!
On Thursday, April 27, 2017 at 3:19:14 PM UTC+5:30, Ankita Chatterjee wrote:I am new to network analysis using Cytoscape. I have names of metabolites and their associated reactions. How to generate a metabolic network in which nodes represent the metabolites and edges represent reactions (So, if two metabolites participate in the same reaction, they should be connected by an edge representing the reaction/enzyme)?
Thank you.
Ankita Chatterjee
Ankita Chatterjee
On 05/01/2017 03:01 AM, Ankita Chatterjee wrote:
I suppose that's technically true, but it's the best we can do. Cytoscape doesn't support hypergraphs.Hi Scooter
I followed the methodology as suggested by you and I could generate a network of the sample file. However, I have few queries: (1) I want the edges to represent reactions. Here, what we are doing is, making the reaction nodes disappear (by assigning node size 1) Thus, edges do not represent the reactions that are common for metabolites.
There are lots of options for moving node labels around. You may one to take a look at the node position options in the Style tab (look under Properties if you don't see it at first).The reaction names are getting overlapped along the edges and looking clumsy. Should it be the correct representation?
No, you can set up a mapping function in the Style tab that will map all nodes with "reac" to size 1.(2) Do we need to assign node size individually to each reaction node manually? In my case the real network will have around 150 metabolites and 400 reactions.
Thanks again for your time!
-- scooter
--On Thursday, April 27, 2017 at 3:19:14 PM UTC+5:30, Ankita Chatterjee wrote:I am new to network analysis using Cytoscape. I have names of metabolites and their associated reactions. How to generate a metabolic network in which nodes represent the metabolites and edges represent reactions (So, if two metabolites participate in the same reaction, they should be connected by an edge representing the reaction/enzyme)?
Thank you.
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Matthias König
you could also simply encode your species-reaction graph in SBML and load the model with cy3sbml.
You have a node attribute type which you can use in your VisualMappings.
The example VisualMappings in cy3sbml make extensive use of it to display the species differently than the reactions.
Let me know if you need help encoding your graph in SBML.
There are very good libraries for that, namely
libsbml (c, c++, python, Java, R bindings)
jsbml (native java library)
The best
Matthias
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Ankita Chatterjee
Ankita Chatterjee
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpdesk+unsubscribe@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpdesk+unsubscribe@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/ecbbe01f-de90-4e33-8e49-199457d57ef9%40googlegroups.com.
Ankita Chatterjee
Ankita Chatterjee
Hello!I am using cytoscape version 3.5.1. After assigning colors to nodes based on their degree, I want to assign different color to some selected nodes. In cytoscape 2.8.3 it was easily achievable (right click on the node and set color), but am unable to do the same in version 3.5.1. Kindly help.Thanks.
National Centre For Cell Sciences, Pune, IndiaSincerelyDST SERB NPDF
Ankita Chatterjee
To unsubscribe from this group and all its topics, send an email to cytoscape-helpdesk+unsubscribe@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
Ankita Chatterjee
Ankita Chatterjee
Ankush Sharma
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGvetfAoJJw0E%3DgBz7ocPnRFc8u58tNQRJMMp3QUDUP7r1LFA%40mail.gmail.com.
Ankita Chatterjee
Ankita Chatterjee
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpdesk+unsubscribe@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGvetfAoJJw0E%3DgBz7ocPnRFc8u58tNQRJMMp3QUDUP7r1LFA%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAALWEk2F45oqpxtggq5N3EaYYj0Jed6Xqz%3DV0moOhyu_nt%2BQvQ%40mail.gmail.com.
Ankita Chatterjee
Ankita Chatterjee
National Centre For Cell Sciences, Pune, IndiaSincerelyDST SERB NPDF
Ankita Chatterjee
On Wed, Jul 26, 2017 at 1:32 PM, Ankush Sharma <ankus...@gmail.com> wrote:
Hi Ankita ,I'm afraid that there is no app in cytoscape to do this. Exporting data and using R or excel could be of use.Best regardsAnkush SharmaBioinformatics Consultant at University of MiamiCASyM (EU-FP7) Postdoctoral Research AssociateInstitute of Clinical Physiology, National Research Council of Italy, Siena ,Italy
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAALWEk2F45oqpxtggq5N3EaYYj0Jed6Xqz%3DV0moOhyu_nt%2BQvQ%40mail.gmail.com.
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpdesk+unsubscribe@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGvetcBvq238g-o%3DBDTpej8uc-NWte8AX%2BEDyg%3DZoHCiz%3DouQ%40mail.gmail.com.
Scooter Morris
clusterMaker2 is certainly compatible with Cytoscape 3.5.1. What version of Java and OS are you on?
-- scooter
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGveteSxDiz89WdSMpiSzy%2B0ttJojMSPxrisPL8Bb2Q4sVNmQ%40mail.gmail.com.
Ankita Chatterjee
Ankita Chatterjee
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGveteSxDiz89WdSMpiSzy%2B0ttJojMSPxrisPL8Bb2Q4sVNmQ%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/2c57c29e-567d-e625-041a-4f39ba687130%40cgl.ucsf.edu.
piet molenaar
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGveteSxDiz89WdSMpiSzy%2B0ttJojMSPxrisPL8Bb2Q4sVNmQ%40mail.gmail.com.
piet...@gmail.com
Department of Oncogenomics, M1-131
Academic Medical Center
University of Amsterdam
Meibergdreef 9
1105 AZ Amsterdam
the Netherlands
tel (+31) 20-5666592
fax (+31) 20-6918626
Ankita Chatterjee
Ankita Chatterjee
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpdesk+unsubscribe@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGveteSxDiz89WdSMpiSzy%2B0ttJojMSPxrisPL8Bb2Q4sVNmQ%40mail.gmail.com.
--Piet Molenaar
piet...@gmail.com
Department of Oncogenomics, M1-131
Academic Medical Center
University of Amsterdam
Meibergdreef 9
1105 AZ Amsterdam
the Netherlands
tel (+31) 20-5666592
fax (+31) 20-6918626
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAHtxcGHUdjW-Mj44MeZPBZp_vj4uXZ0LmKLnw65Z4OYfxOdXOQ%40mail.gmail.com.
piet molenaar
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAHtxcGHUdjW-Mj44MeZPBZp_vj4uXZ0LmKLnw65Z4OYfxOdXOQ%40mail.gmail.com.
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGvetczJ_WRDSLzViWOAgSQU9BFXp-AX02Rhe%2BiBjsF_oW%2BvA%40mail.gmail.com.
Barry Demchak
Hi, all –
I don’t know what is up with this … I just tried this on my PC and had no problems of any sort.
Regardless, while we consider what could be going wrong, I’d like to make a copy of ClusterMaker2 available to Ankita here: http://chianti.ucsd.edu/~bdemchak/clusterMaker2-1.1.0.jar
Ankita … does this happen on any other PC? More information is helpful.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/ecbbe01f-de90-4e33-8e49-199457d57ef9%40googlegroups.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGvetfAoJJw0E%3DgBz7ocPnRFc8u58tNQRJMMp3QUDUP7r1LFA%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAALWEk2F45oqpxtggq5N3EaYYj0Jed6Xqz%3DV0moOhyu_nt%2BQvQ%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGvetcBvq238g-o%3DBDTpej8uc-NWte8AX%2BEDyg%3DZoHCiz%3DouQ%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGveteSxDiz89WdSMpiSzy%2B0ttJojMSPxrisPL8Bb2Q4sVNmQ%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
--
Piet Molenaar
piet...@gmail.com
Department of Oncogenomics, M1-131
Academic Medical Center
University of Amsterdam
Meibergdreef 9
1105 AZ Amsterdam
the Netherlands
tel (+31) 20-5666592
fax (+31) 20-6918626
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAHtxcGHUdjW-Mj44MeZPBZp_vj4uXZ0LmKLnw65Z4OYfxOdXOQ%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGvetczJ_WRDSLzViWOAgSQU9BFXp-AX02Rhe%2BiBjsF_oW%2BvA%40mail.gmail.com.
Barry Demchak
Hi, Ankita –
Something’s wrong here … Windows 7/64 is OK for Cytoscape 3.5.1, but Java 7 is not. Cytoscape shouldn’t even start without Java 8. Would you mind verifying your Java version … on the Cytoscape splash screen (during startup) or on the Help | About panel.
Thanks.
From: cytoscape...@googlegroups.com [mailto:cytoscape...@googlegroups.com] On Behalf Of Ankita Chatterjee
Sent: Wednesday, August 09, 2017 11:06 PM
To: cytoscape...@googlegroups.com
Subject: Re: [cytoscape-helpdesk] Re: Generating metabolic network
Hi Scooter,
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/ecbbe01f-de90-4e33-8e49-199457d57ef9%40googlegroups.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGvetfAoJJw0E%3DgBz7ocPnRFc8u58tNQRJMMp3QUDUP7r1LFA%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAALWEk2F45oqpxtggq5N3EaYYj0Jed6Xqz%3DV0moOhyu_nt%2BQvQ%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGvetcBvq238g-o%3DBDTpej8uc-NWte8AX%2BEDyg%3DZoHCiz%3DouQ%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGveteSxDiz89WdSMpiSzy%2B0ttJojMSPxrisPL8Bb2Q4sVNmQ%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/2c57c29e-567d-e625-041a-4f39ba687130%40cgl.ucsf.edu.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGvetdyqk9vq%2BdZEdXjz8OBopS-PCFRvPFshnJQNd9ALE%2B56Q%40mail.gmail.com.
Ankita Chatterjee
Ankita Chatterjee
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/ecbbe01f-de90-4e33-8e49-199457d57ef9%40googlegroups.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGvetfAoJJw0E%3DgBz7ocPnRFc8u58tNQRJMMp3QUDUP7r1LFA%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAALWEk2F45oqpxtggq5N3EaYYj0Jed6Xqz%3DV0moOhyu_nt%2BQvQ%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGvetcBvq238g-o%3DBDTpej8uc-NWte8AX%2BEDyg%3DZoHCiz%3DouQ%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGveteSxDiz89WdSMpiSzy%2B0ttJojMSPxrisPL8Bb2Q4sVNmQ%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/2c57c29e-567d-e625-041a-4f39ba687130%40cgl.ucsf.edu.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this group and stop receiving emails from it, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/CAPGvetdyqk9vq%2BdZEdXjz8OBopS-PCFRvPFshnJQNd9ALE%2B56Q%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/JnjUherjREU/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
To post to this group, send email to cytoscape-helpdesk@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/011901d311ee%248c6cde30%24a5469a90%24%40gmail.com.
