1) Background gene set for ClueGO and 2) summary statistics (p-value and FDR)
537 views
Skip to first unread message
BIOMICS
Feb 14, 2018, 1:45:06 AM2/14/18
to cytoscape-helpdesk
Dear all,
1. While using ClueGO in Cytoscape is it possible to load custom background gene list while doing any form of analysis (pathway, biological process etc) using ClueGO analysis.
2. After deriving a network using ClueGO will it be possible to derive (a) p-value and (b) FDR for each node and edges as well as for each gene or compounds that were used to create the network.
3. Will it be possible to get a summary statistics for all nodes, edges and input genes or metabolites associated with the network?
Please let me know.
Thanks, Nara
Alex Pico
Feb 22, 2018, 2:23:26 PM2/22/18
to cytoscape-helpdesk
Forwarded to ClueGO app author listed here: http://apps.cytoscape.org/apps/cluego
Bernhard
Feb 25, 2018, 9:35:34 AM2/25/18
to cytoscape-helpdesk
Hi Nara,
On Wednesday, February 14, 2018 at 7:45:06 AM UTC+1, BIOMICS wrote:
Dear all,1. While using ClueGO in Cytoscape is it possible to load custom background gene list while doing any form of analysis (pathway, biological process etc) using ClueGO analysis.
You can load data sets under CluePedia and make correlation matrices, or ttests, or visualize the expression data on gene nodes in the network.
2. After deriving a network using ClueGO will it be possible to derive (a) p-value and (b) FDR for each node and edges as well as for each gene or compounds that were used to create the network.
ClueGO only calculates enrichment (fisher's exact test) p-values for the GO / Pathway nodes.
3. Will it be possible to get a summary statistics for all nodes, edges and input genes or metabolites associated with the network?
What kind of statistics are you thinking on? Normally enrichment analysis stats are only for Pathway nodes.
Please let me know.Thanks, Nara
Best
Narasimhan Kothandaraman
Feb 25, 2018, 9:49:26 AM2/25/18
to cytoscape...@googlegroups.com
Hi Bernhard,
I) Can you please share the documentation on how to create 1) correlation matrices, 2) ttests, 3) visualize the expression data on gene nodes in the network.
II) Also please let me know how to get enrichment analysis stats for Pathway nodes.
Thank you,
Nara
From: Bernhard <gabriel...@gmail.com>
To: cytoscape-helpdesk <cytoscape...@googlegroups.com>
Sent: Sunday, 25 February 2018, 22:35
Subject: [cytoscape-helpdesk] Re: 1) Background gene set for ClueGO and 2) summary statistics (p-value and FDR)
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/Bxt9jfJetdk/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/1a0c24e0-d262-4d56-a6fc-8d46b785933e%40googlegroups.com.
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/Bxt9jfJetdk/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/1a0c24e0-d262-4d56-a6fc-8d46b785933e%40googlegroups.com.
Bernhard
Mar 1, 2018, 1:45:01 PM3/1/18
to cytoscape-helpdesk
Hi Nara,
On Sunday, February 25, 2018 at 3:49:26 PM UTC+1, BIOMICS wrote:
Hi Bernhard,I) Can you please share the documentation on how to create 1) correlation matrices, 2) ttests, 3) visualize the expression data on gene nodes in the network.
We are working on this, but we don't have much time at the moment, so it will still take a moment.
II) Also please let me know how to get enrichment analysis stats for Pathway nodes.
What stats do you mean? Normally you have all the pvalues and results in the ClueGO result panel after you run the analysis
Thank you,Nara
Best
From: Bernhard <gabriel...@gmail.com>
To: cytoscape-helpdesk <cytoscape...@googlegroups.com>
Sent: Sunday, 25 February 2018, 22:35
Subject: [cytoscape-helpdesk] Re: 1) Background gene set for ClueGO and 2) summary statistics (p-value and FDR)
Hi Nara,--
On Wednesday, February 14, 2018 at 7:45:06 AM UTC+1, BIOMICS wrote:Dear all,1. While using ClueGO in Cytoscape is it possible to load custom background gene list while doing any form of analysis (pathway, biological process etc) using ClueGO analysis.You can load data sets under CluePedia and make correlation matrices, or ttests, or visualize the expression data on gene nodes in the network.2. After deriving a network using ClueGO will it be possible to derive (a) p-value and (b) FDR for each node and edges as well as for each gene or compounds that were used to create the network.ClueGO only calculates enrichment (fisher's exact test) p-values for the GO / Pathway nodes.3. Will it be possible to get a summary statistics for all nodes, edges and input genes or metabolites associated with the network?What kind of statistics are you thinking on? Normally enrichment analysis stats are only for Pathway nodes.Please let me know.Thanks, NaraBest
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/Bxt9jfJetdk/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
BIOMICS
Mar 14, 2018, 10:42:07 PM3/14/18
to cytoscape-helpdesk
Hi Bernhard,
In CLueGO what is the default background gene set on which the networks are created?
in the Reference Set Options in CLueGO there are three options:
1) Selected Ontologies Reference Set
2) Predefined IDs reference set
3) Custom reference set.
Can you please explain item (1) and (2). When we say Selected Ontologies Reference Set, what will be the background genes that ClueGO will be using to create the network?
Thank you,
Nara
Narasimhan Kothandaraman
Mar 15, 2018, 10:30:12 PM3/15/18
to cytoscape...@googlegroups.com
Hi Bernhard,
In ClueGO documentation I can see a results folder on page number 17. http://www.ici.upmc.fr/cluego/ClueGODocumentation.pdf
Please let me know if there is an option in ClueGO to create this by default settings or specific steps to create this.
Regarding Kappa, score matrix is it possible for Cytoscape to generate this. The documentation mentions the use of Genesis. Is it a third party software or does Cytoscape can create this Kappa score matric by default.
Please clarify.
Many thanks
Nara
From: BIOMICS <obg...@gmail.com>
To: cytoscape-helpdesk <cytoscape...@googlegroups.com>
Sent: Thursday, 15 March 2018, 10:42
Subject: [cytoscape-helpdesk] Re: 1) Background gene set for ClueGO and 2) summary statistics (p-value and FDR)
--
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/Bxt9jfJetdk/unsubscribe.
You received this message because you are subscribed to a topic in the Google Groups "cytoscape-helpdesk" group.
To unsubscribe from this topic, visit https://groups.google.com/d/topic/cytoscape-helpdesk/Bxt9jfJetdk/unsubscribe.
To unsubscribe from this group and all its topics, send an email to cytoscape-helpd...@googlegroups.com.
To post to this group, send email to cytoscape...@googlegroups.com.
Visit this group at https://groups.google.com/group/cytoscape-helpdesk.
To view this discussion on the web visit https://groups.google.com/d/msgid/cytoscape-helpdesk/489c752a-52e7-48ac-9f4c-21da85778361%40googlegroups.com.
Bernhard
Mar 22, 2018, 6:20:48 AM3/22/18
to cytoscape-helpdesk
Hi Nara, to answer first your questions here. The reference set is used to calculate the enrichment statistics. You can find more info how it is calculated directly if you click on the question mark icons.
1) Selected Ontologies Reference Set:
This reference gene set is used by default and uses all genes found in the selected ontologies as reference set, so the sum of all unique genes found.
1) Selected Ontologies Reference Set:
This reference gene set is used by default and uses all genes found in the selected ontologies as reference set, so the sum of all unique genes found.
2) Predefined IDs reference set
Here you can select a predefined reference set that you can add as annotation file. So if e.g. you analyze affymetrix data you could select all genes on the affymetrix chip as reference.
3) Custom reference set.
3) Custom reference set.
Here you can upload what ever list of genes you want to be the reference set. All the ontologies will then be imputed to this reference set.
I hope it is more clear now
Bernhard
Mar 22, 2018, 6:31:07 AM3/22/18
to cytoscape-helpdesk
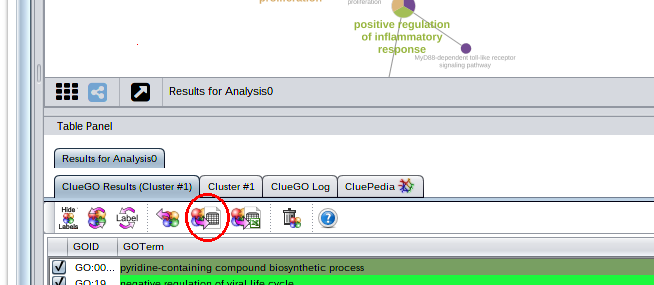
Hi Nara, the binary term-gene matrix and the term-term matrix with the kappascore values is generated when ever you make and enrichment analysis. You have to save the data (see down marked in red). This will give you the tab delimited files with all the corresponding values that you can open then with e.g. excel or libreoffice. To generate heatmaps you can use any clustering program like e.g. Genesis that you can download here http://genome.tugraz.at/genesisclient/genesisclient_description.shtml.
Best
Best
To unsubscribe from this group and all its topics, send an email to cytoscape-helpdesk+unsub...@googlegroups.com.
Reply all
Reply to author
Forward
0 new messages
