How to use DICOMDIR format
86 views
Skip to first unread message
刘政
May 25, 2021, 11:25:33 PM5/25/21
to TVB Users
Hi
The DTI data I've got is from Siemens 3T MRI scanner, and the data it gives me is in DICOMDIR format, which contains a lot of series. I used A software called MiscroDICOM to open it, and there are several series,
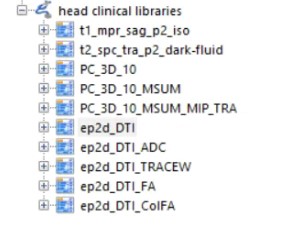

Does anyone know which one I should use for VEP?
Thank you,
Barry
WOODMAN Michael
May 26, 2021, 2:09:05 AM5/26/21
to tvb-...@googlegroups.com
Hi
You would use the t1_mpr (and maybe the t2_spc) for FreeSurfer and the ep2d_DTI for the diffusion part. The important step is to check the sequence contains the gradient information, which can be done with mrinfo command from MRtrix3.
cheers,
Marmaduke
From: tvb-...@googlegroups.com <tvb-...@googlegroups.com> on behalf of 刘政 <lz02...@gmail.com>
Sent: Wednesday, May 26, 2021 5:25:33 AM
To: TVB Users
Subject: [TVB] How to use DICOMDIR format
Sent: Wednesday, May 26, 2021 5:25:33 AM
To: TVB Users
Subject: [TVB] How to use DICOMDIR format
--
You received this message because you are subscribed to the Google Groups "TVB Users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to tvb-users+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/tvb-users/4f8244eb-9e8c-42d0-9255-bff561ed6f9an%40googlegroups.com.
You received this message because you are subscribed to the Google Groups "TVB Users" group.
To unsubscribe from this group and stop receiving emails from it, send an email to tvb-users+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/tvb-users/4f8244eb-9e8c-42d0-9255-bff561ed6f9an%40googlegroups.com.
刘政
May 26, 2021, 4:00:38 AM5/26/21
to TVB Users
Hi
When you say gradient information, does itmena like this. When I open the DTI of ep2d_DTI series, there are 62 figures with 1024 * 2014 size, and in each figure, there're 8*8=64 brains in it. Like this
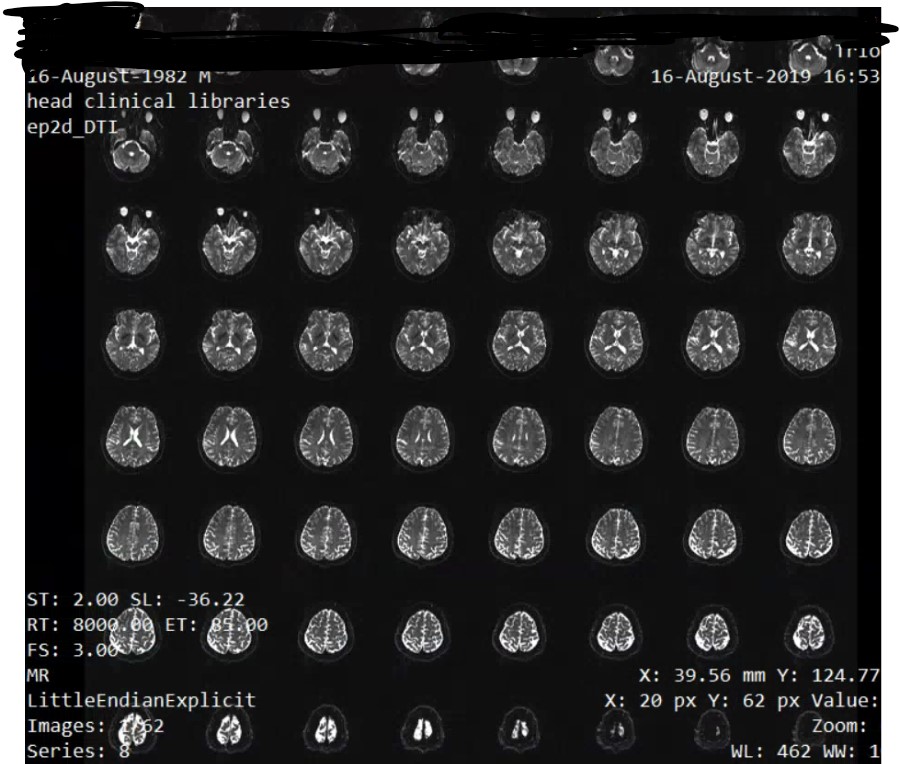
And do you know some tools that would help me trnasfer the DICOMDIR file to DICOM format, the software I am using now can do this, but if I use mrinfo to see the info of the dcm file transfered from the MiscroDICOM viewer, it gives me error like this
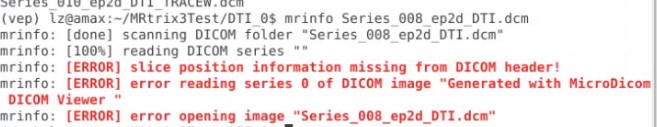
Thank you
Barry
WOODMAN Michael
May 26, 2021, 4:58:02 AM5/26/21
to tvb-...@googlegroups.com
hi
This would be the right series, but if MRtrix3 cannot read it, then you would need to find a converter to a format that it can read, perhaps nifti?
I can't help further, sorry, and please refrain from posting personal image data to the mailing list.
cheers,
Marmaduke
Sent: Wednesday, May 26, 2021 10:00:37 AM
To: TVB Users
Subject: Re: [TVB] How to use DICOMDIR format
To: TVB Users
Subject: Re: [TVB] How to use DICOMDIR format
To view this discussion on the web visit
https://groups.google.com/d/msgid/tvb-users/2d22295b-9b46-4501-8957-95c971588a51n%40googlegroups.com.
刘政
May 26, 2021, 7:31:58 AM5/26/21
to TVB Users
Hi
Got it! And I just converted that series to nii.gz format, and use mrinfo command, get this
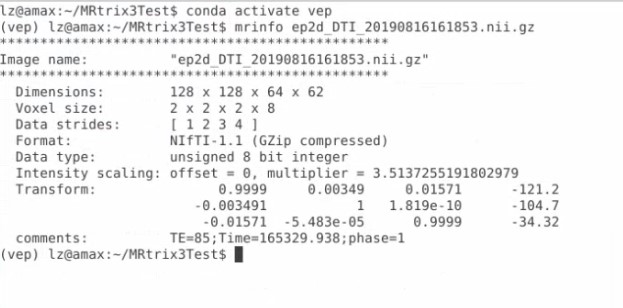
Is this the right output?
Thank you
Barry
WOODMAN Michael
May 26, 2021, 3:22:26 PM5/26/21
to tvb-...@googlegroups.com
Hi
That looks good except you will need the gradient information in addition to the nifti image. Frequently this is provided in a FSL convention with two files bvecs and bvals. lease refer to MRtrix documentation for more information .
Cheers
Marmaduke
> On 26 May 2021, at 13:32, 刘政 <lz02...@gmail.com> wrote:
>
>
That looks good except you will need the gradient information in addition to the nifti image. Frequently this is provided in a FSL convention with two files bvecs and bvals. lease refer to MRtrix documentation for more information .
Cheers
Marmaduke
> On 26 May 2021, at 13:32, 刘政 <lz02...@gmail.com> wrote:
>
>
> Hi
>
> Got it! And I just converted that series to nii.gz format, and use mrinfo command, get this
>
> Got it! And I just converted that series to nii.gz format, and use mrinfo command, get this
刘政
Jun 8, 2021, 5:50:21 AM6/8/21
to TVB Users
Hi
May I ask that how do you guys get the weight.txt, aka the weight matrix, with T1 MRI and DTI(T1 is a .nii.gz file, and DTI consisting of .nii.gz bvec bval)? I saw two repositories here https://github.com/the-virtual-brain/tvb-recon and https://github.com/ins-amu/scripts/tree/v0.4-CH. They all look working. And as to the Scripts, it has many tags, any recommendation?
Thank you
Barry
刘政
Jun 8, 2021, 6:24:36 AM6/8/21
to TVB Users
Hi
And is the parcellation optional, for I notice a new atlas called VEP-atlas
Thank you
Barry
刘政
Jun 8, 2021, 11:13:49 AM6/8/21
to TVB Users
Hi all
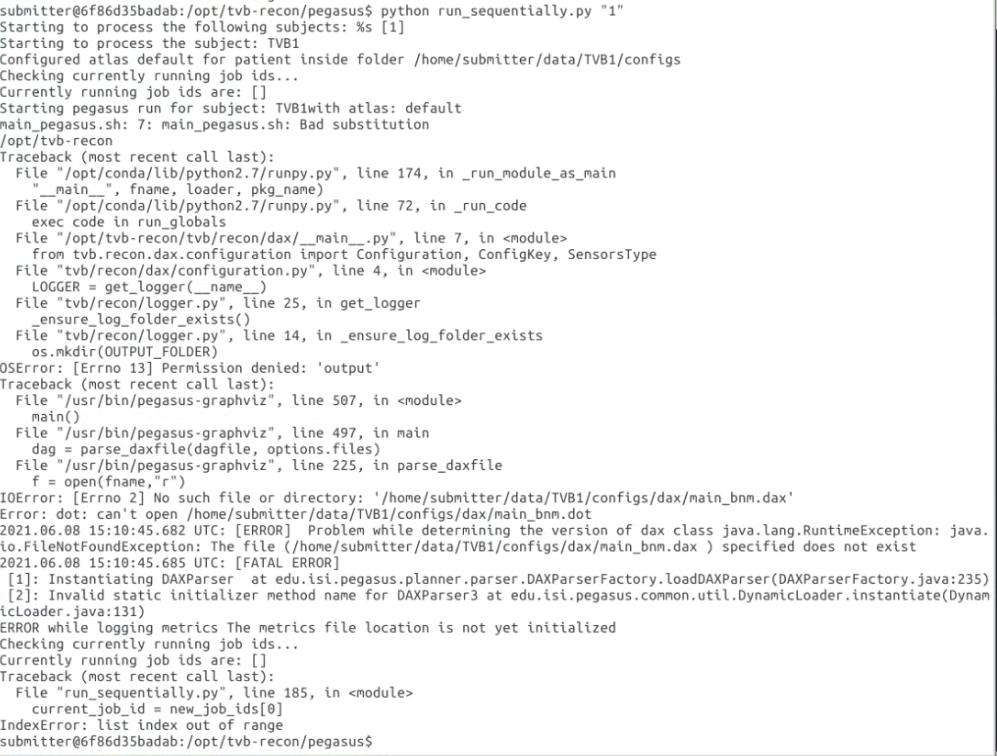
I am using tvb-recon(https://github.com/the-virtual-brain/tvb-recon) docker image to get the weight.txt of structural
construction.
And I met this error:

Reply all
Reply to author
Forward
0 new messages
