Libra quantification on TMT 16-plex returns zeros only
Yasir Ahmed
Luis Mendoza
--
You received this message because you are subscribed to the Google Groups "spctools-discuss" group.
To unsubscribe from this group and stop receiving emails from it, send an email to spctools-discu...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/7cb42706-53ae-4c94-acd2-ea0293fea8b2n%40googlegroups.com.
Yasir Ahmed
Can you try re-running Libra without the centroiding option? (i.e. set type="0") Or are your mzML data not centroided?
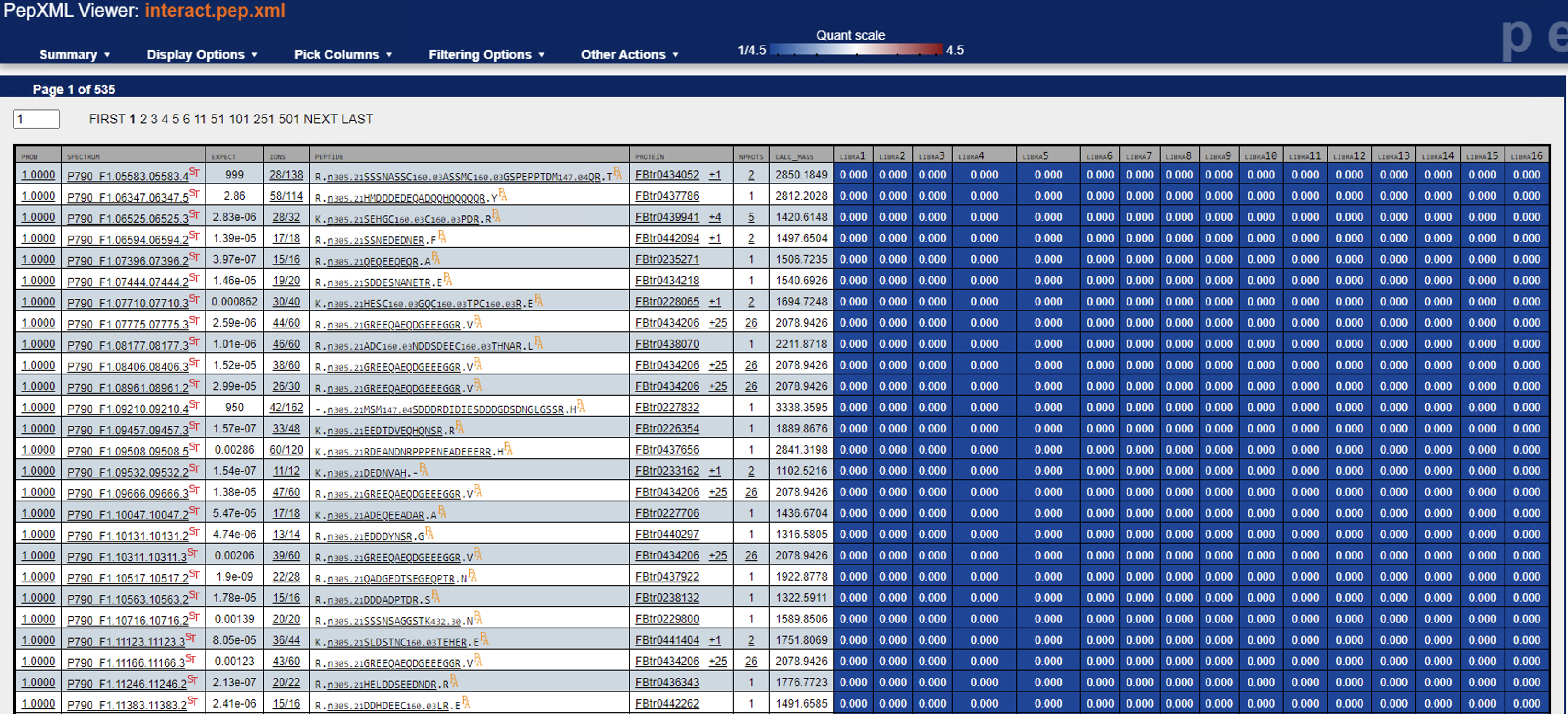
Another thing to try: are you able to open the PeptideProphet + Libra results in the PepXMLViewer? You should be able to see if there are a significant number of quantified PSMs -- you should also be able to examine a few individual spectra and zoom in on the reporter ion signals.
Lastly, what version of TPP are you running?
Cheers,--LuisOn Thu, Sep 8, 2022 at 12:02 PM Yasir Ahmed <yaza...@gmail.com> wrote:Greetings,I'm having difficulty performing TMT quantification using Libra on a 16-plex dataset. Basically, the "quantification.tsv" file that Libra outputs only contains zeros for both peptides and proteins. The Comet search runs just fine. I'm at a loss for where things are going wrong. The Mass Spec facility used ProteomeDiscoverer for the quantification, and it worked fine so the issue is not the raw data.My TPP run repository can be found here, and the "condition.xml" file for Libra—along with the Comet params file—is in the "params" folder. The commands used to run the TPP processes can be found in the file called "Snakefile". I'm happy to provide any additional information needed to help troubleshoot this. Any pointers/advise would be greatly appreciated.Cheers,Yasir--
You received this message because you are subscribed to the Google Groups "spctools-discuss" group.
To unsubscribe from this group and stop receiving emails from it, send an email to spctools-discu...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/7cb42706-53ae-4c94-acd2-ea0293fea8b2n%40googlegroups.com.
--
You received this message because you are subscribed to the Google Groups "spctools-discuss" group.
To unsubscribe from this group and stop receiving emails from it, send an email to spctools-discu...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/CACyS9bo%3DQxLBhn3H%2B7cg9pTKXu1Yp1bTLwb%2BfF%3DR4y%2BujoF%3DxQ%40mail.gmail.com.
Yasir Ahmed
On Sep 8, 2022, at 5:07 PM, Yasir Ahmed <yaza...@gmail.com> wrote:
Hi Luis,Thanks a lot for the response.
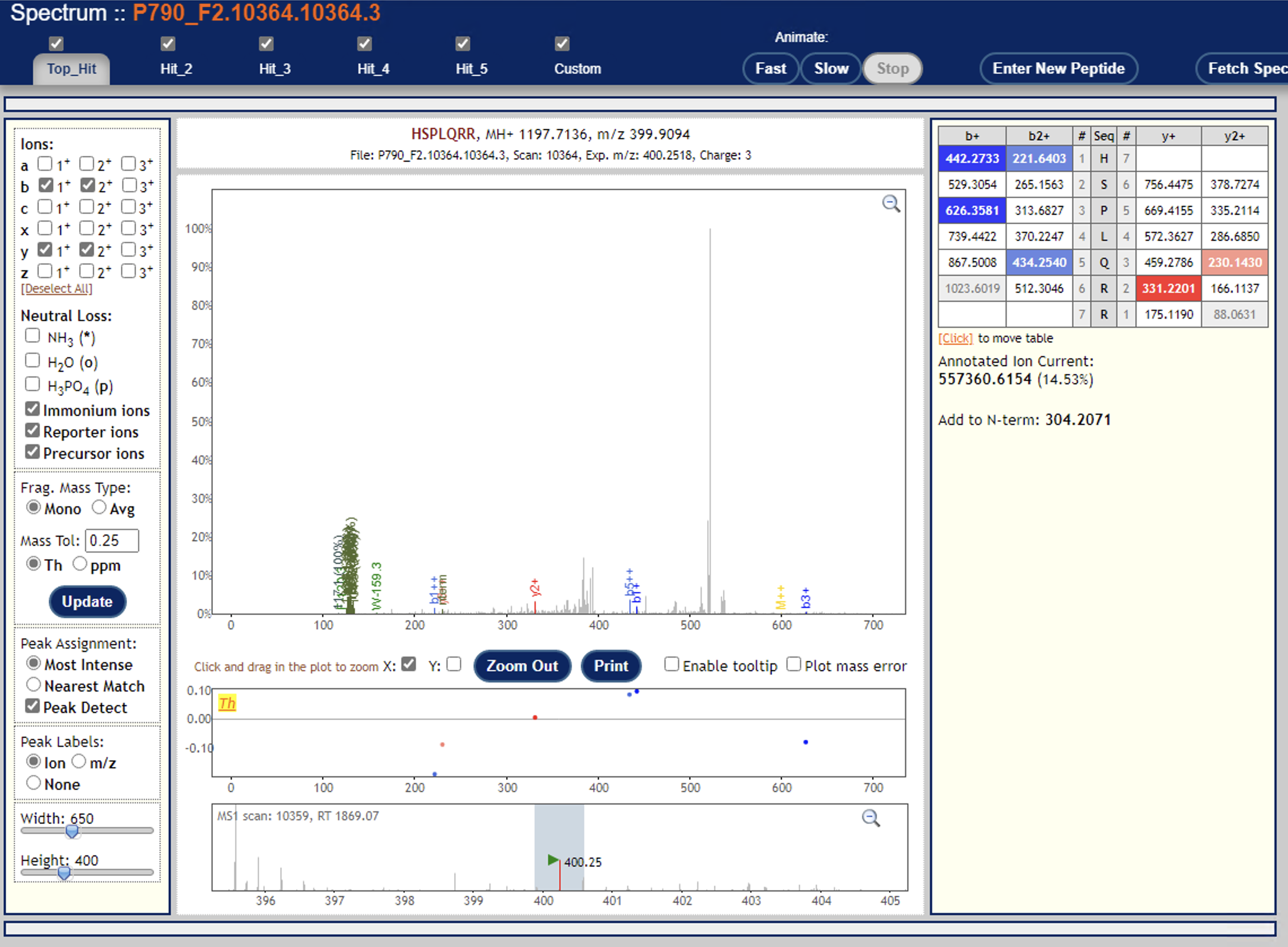
Can you try re-running Libra without the centroiding option? (i.e. set type="0") Or are your mzML data not centroided?I just tried that, but no difference in outcome.Another thing to try: are you able to open the PeptideProphet + Libra results in the PepXMLViewer? You should be able to see if there are a significant number of quantified PSMs -- you should also be able to examine a few individual spectra and zoom in on the reporter ion signals.I’m able to look at a few individual spectra, and I think they all look mostly good—but frankly I’m not too sure how to evaluate their suitability (see example screenshots below).Lastly, what version of TPP are you running?6.1.0Thanks again!Yasir
<Screen Shot 2022-09-08 at 5.00.50 PM.png><Screen Shot 2022-09-08 at 5.01.42 PM.png>
Eric Deutsch
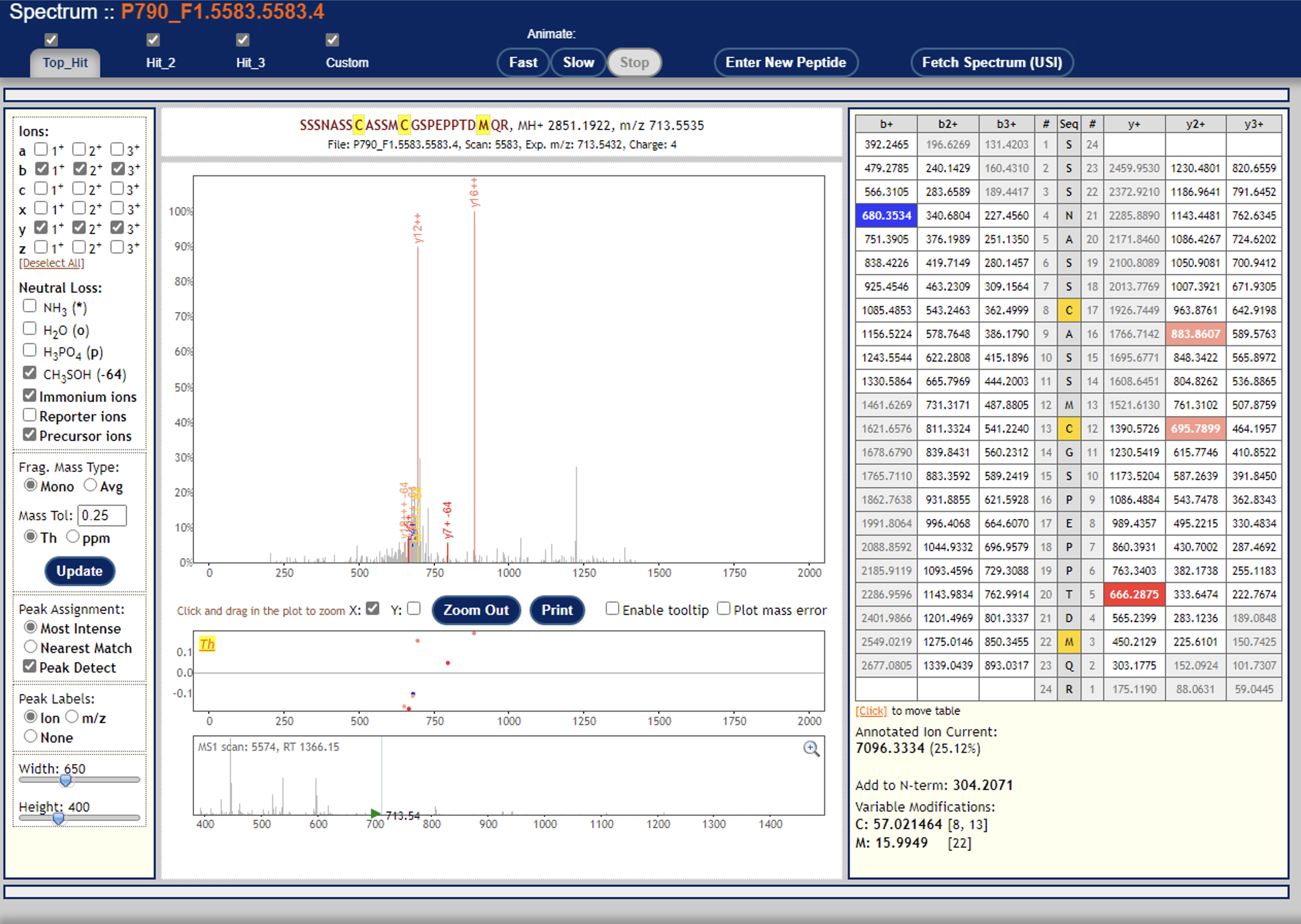
I don’t see any signal down where the reporter ions are at ~120 m/z
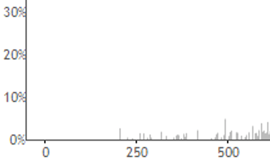
In fact, that looks a lot like an ion trap spectrum, which typically don’t have signal down at ~120.
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/B61146FA-374D-4D4C-8453-C8F388541B27%40gmail.com.
Eric Deutsch
I’m thinking that ion trap spectra are usually unsuitable for TMT:
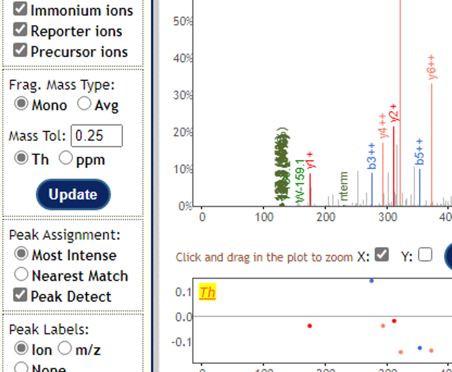
I think you would want to acquire fragmentation spectra with Orbitrap HCD?
From: spctools...@googlegroups.com <spctools...@googlegroups.com> On Behalf Of Yasir Ahmed
Sent: Thursday, September 8, 2022 2:19 PM
To: spctools...@googlegroups.com
Subject: Re: [spctools-discuss] Libra quantification on TMT 16-plex returns zeros only
Hi Luis,
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/5C533052-EF37-4516-AE3D-D90A364E5E68%40gmail.com.
Luis Mendoza
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/aad015ab22f19ee0cea8cd2cdc24627d%40mail.gmail.com.
Yasir Ahmed
On Sep 8, 2022, at 6:15 PM, 'Luis Mendoza' via spctools-discuss <spctools...@googlegroups.com> wrote:
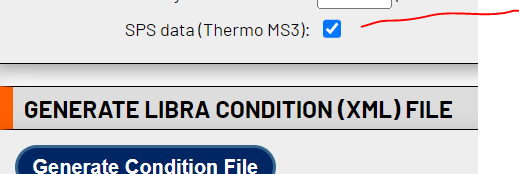
Hi Yasir,Is your data Thermo SPS where the reporter ions are in MS3? If so, please check the relevant box when producing the condition file:
<image.png>As Eric noted, it does not seem like your MS2 spectra contain reporter ion signals, at least from the one spectrum you posted. Are there others where this is not the case?Cheers,--Luis
On Thu, Sep 8, 2022 at 2:28 PM 'Eric Deutsch' via spctools-discuss <spctools...@googlegroups.com> wrote:
I’m thinking that ion trap spectra are usually unsuitable for TMT:
<image001.png>
I think you would want to acquire fragmentation spectra with Orbitrap HCD?
From: spctools...@googlegroups.com <spctools...@googlegroups.com> On Behalf Of Yasir Ahmed
Sent: Thursday, September 8, 2022 2:19 PM
To: spctools...@googlegroups.com
Subject: Re: [spctools-discuss] Libra quantification on TMT 16-plex returns zeros onlyHi Luis,
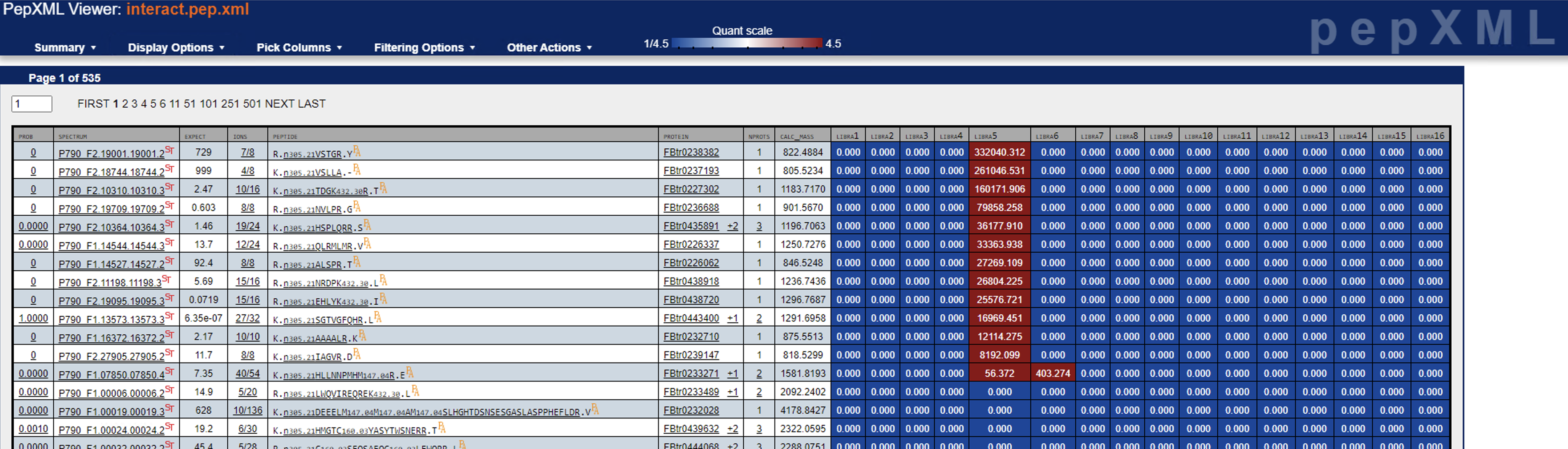
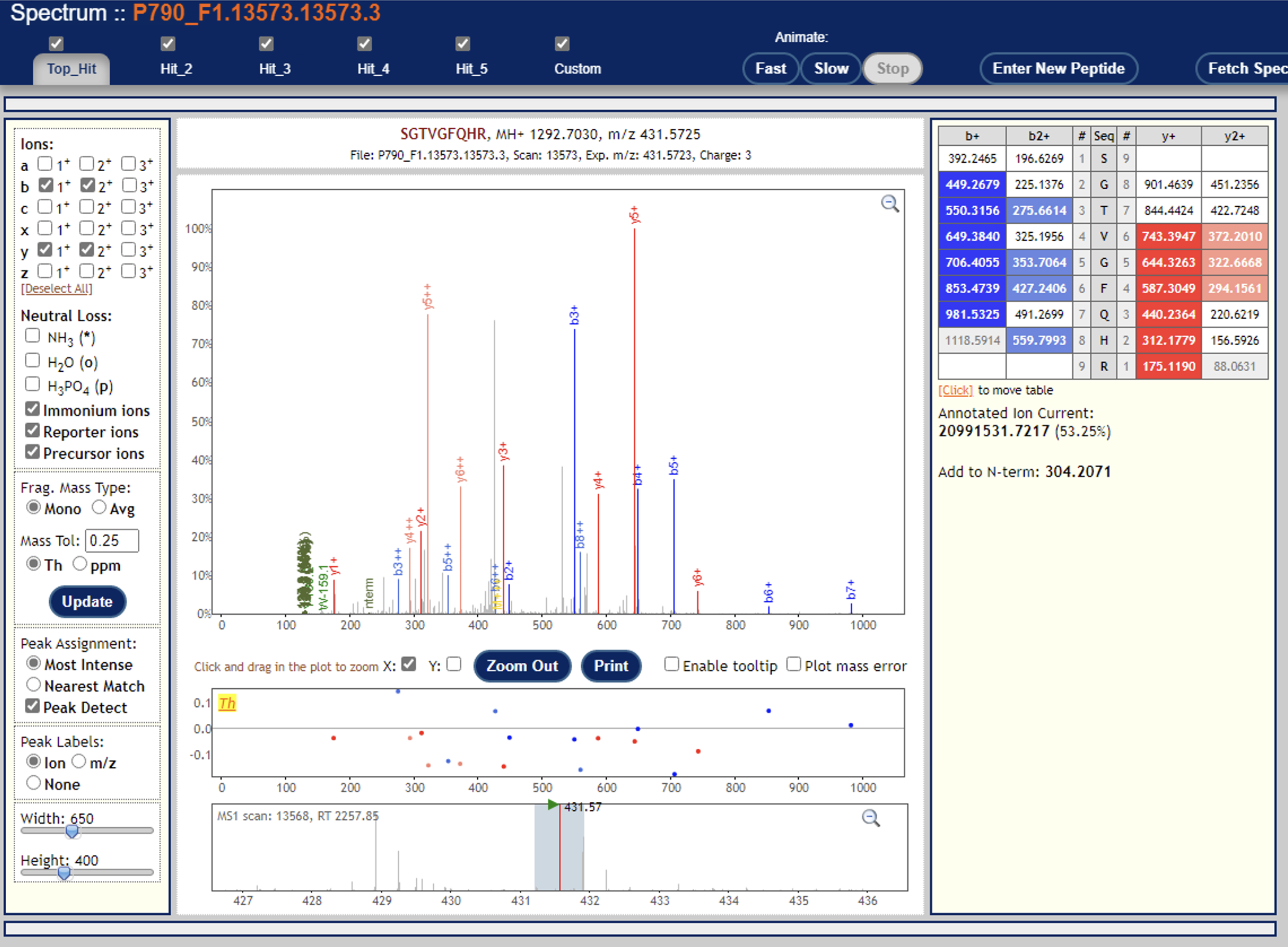
Just to follow up: I found a handful of cases where one of the channels has super high quantification and observable reporter ions (see screenshots below), albeit it should be way more than this, I believe. I’ll keep digging, but let me know if you have any other ideas.
Again, thanks for you suggestions.
Cheers,
Yasir
<image002.png>
<image003.png>
<image004.png>
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/CACyS9boObJ2nFUNDd_1K%2BMghDJy62GQEYu4R6zRn2CuS-7JwBQ%40mail.gmail.com.
Luis Mendoza
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/7EBE121E-E4BC-495C-B288-EE9B84CA78F9%40gmail.com.
