Changes in number of peptides identifications in TPP 6.0 RC
53 views
Skip to first unread message
Oded
Apr 12, 2021, 2:49:16 PM4/12/21
to spctools-discuss
Hi there,
We recently download the TPP 6.0 RC and while using it we noticed that we obtain fewer peptides IDs than what we got for the same dataset and search output with version 5.2.
Many of the missing peptides seem to have decent Expect value and MS/MS following a visual inspection.
This seems that this is due to changes in PeptideProphet and are not related to the search itself that was done with Comet.
Is there any additional parameter that should be included in the new version in order to restore the missing peptides?
Many thanks,
This seems that this is due to changes in PeptideProphet and are not related to the search itself that was done with Comet.
Is there any additional parameter that should be included in the new version in order to restore the missing peptides?
Many thanks,
Oded
David Shteynberg
Apr 12, 2021, 3:17:53 PM4/12/21
to spctools-discuss
Hi Oded, Which release candidate are you referring to? The earlier candidates may have a bug that is corrected in a later version. If you can share some data and specifics about the missing PSMs I can run it here and troubleshoot the problem.
Thanks!
David
P.S. I am on vacation this week so will troubleshoot next week.
--
You received this message because you are subscribed to the Google Groups "spctools-discuss" group.
To unsubscribe from this group and stop receiving emails from it, send an email to spctools-discu...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/b7c2e3b7-3061-4ec7-94d8-f70e43538f2an%40googlegroups.com.
Oded
Apr 12, 2021, 4:38:27 PM4/12/21
to spctools-discuss
Hi David,
Thank you for the quick reply. We are using TPP v6.0.0-rc14 Noctilucent, Build 202103031119-8400 (Windows_NT-x86_64).
As for the data let me know when you are back and I will transfer you either the raw file or the search results.
Enjoy your vacation.
Thanks,
Oded
David Shteynberg
Apr 12, 2021, 4:46:08 PM4/12/21
to spctools-discuss
I don't mind taking a crack at it over vacation. Please let me know where I can pull the data from. I might not have quick solution for you but I can get started looking for the problem. Which search engine did you use here? I would need mzML data search results and the fasta database to get started.
Thanks!
Thanks!
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/7f5e47e5-56df-4552-88ca-b6d4396f3d8en%40googlegroups.com.
Oded
Apr 12, 2021, 5:09:05 PM4/12/21
to spctools-discuss
Hi David,
You can find the mzML, Comet parameters and FASTA file here:https://www.dropbox.com/t/Yld0ZBWhsuFajeLj
The link is valid for 7 days.
Many thanks,
Oded
Oded
Apr 12, 2021, 5:15:00 PM4/12/21
to spctools-discuss
And also the search results with the new vs the old version (although I think they are more or less the same): https://www.dropbox.com/t/8VyBVdeuoWdk982q
David Shteynberg
Apr 12, 2021, 8:38:33 PM4/12/21
to spctools-discuss
Dear Oded,
Thanks for this. I ran a quick test and I actually observed a few more PSMs for PeptideProphet 6.0.0-rc14 for the same PeptideProphet probability cutoff for this dataset. Is the issue you see with PeptideProphet or iProphet results? Which spectra were getting excluded in the your testing of 5.2.0 vs 6.0.0, can you give me some specific examples? Are you certain you are using identical analysis parameters for PeptideProphet and what are those parameters?
Cheers,
David
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/3baf2dd3-f237-4c9d-82a2-54d563ccf5e6n%40googlegroups.com.
Oded
Apr 13, 2021, 5:07:11 PM4/13/21
to spctools-discuss
Dear David,
This is PeptideProphet issue (we don't use iProphet for this analysis).
With the search parameters that I sent you followed by running the PeptideProphet command of xinteract -Ninteract.pep.xml -p0.05 -l7 -PPM -OANEp -dDECOY Seq69478_QE2.pep.xml,
With the search parameters that I sent you followed by running the PeptideProphet command of xinteract -Ninteract.pep.xml -p0.05 -l7 -PPM -OANEp -dDECOY Seq69478_QE2.pep.xml,
and lastly using a cutoff of 1% error rate we obtain with TPP 5.2 2836 correct assignments and with TPP 6.0 only 2228 correct assignments. The probability for 1% error rate is 0.84 in TPP 5.2 and 0.9090 in TPP 6.0).
In the following link, you will find the interact files (new and old):https://www.dropbox.com/t/mwtpCLeEJRLCwbi1
In the following link, you will find the interact files (new and old):https://www.dropbox.com/t/mwtpCLeEJRLCwbi1
Thanks,
Oded
David Shteynberg
Apr 14, 2021, 1:34:42 AM4/14/21
to spctools-discuss
Hi Oded,
I think the old 5.2.0 might be under estimating the error rate as compared to the new release candidate. You cannot see the decoys here because of the settings you used. However your database has two independent sets of decoys available DECOY0 and DECOY1. Can you use one of the set for PeptideProphet and use the other set to get a decoy count at a given PeptideProphet probability cutoff. I suspect the 5.2.0 TPP will show more unknown decoys than the new release candidate. Can you test if this is the case here? I can run a more in depth analysis when I am back from vacation next week.
Cheers,
David
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/3030a2db-c902-4cb4-837d-20f385662a18n%40googlegroups.com.
Oded
Apr 14, 2021, 5:18:09 AM4/14/21
to spctools-discuss
Hi David,
This time I used xinteract -NinteractDecoyCount.pep.xml -p0.05 -l7 -PPM -OANEdp -dDECOY0 Seq69478_QE2.pep.xml.
With TPP 5.2 the count of DECOY1 ids at 1% error rate is 84 and the total count of DECOY ids is 151 (out of 7102 PSMs).
With TPP 6.0 the count of DECOY1 ids at 1% error rate is 7 and the total count of DECOY ids is 9 (out of 5308 PSMs).
You can find the new interact files here:https://www.dropbox.com/t/s7ZqgkP1f0Yzl0Xn.
Many thanks,
Oded
David Shteynberg
Apr 14, 2021, 1:28:29 PM4/14/21
to spctools-discuss
Hi Oded,
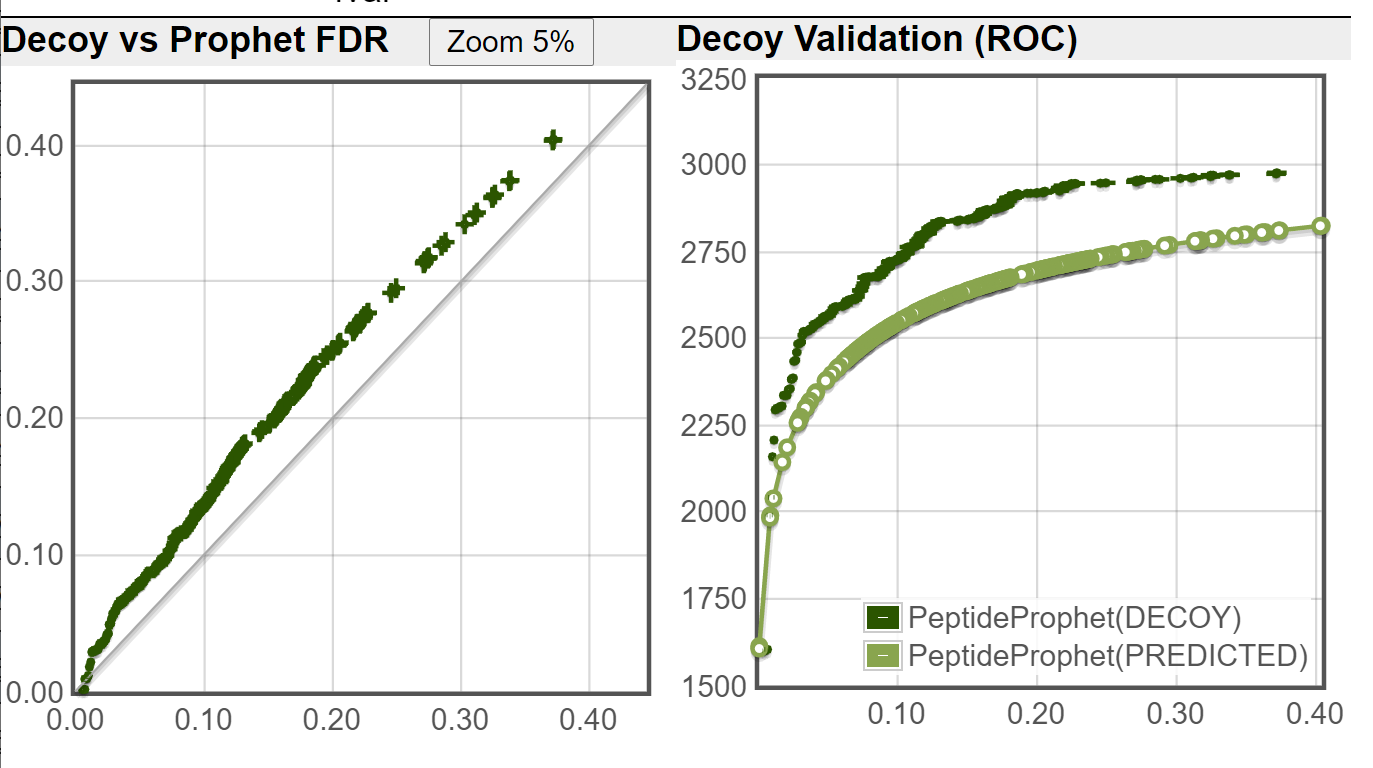
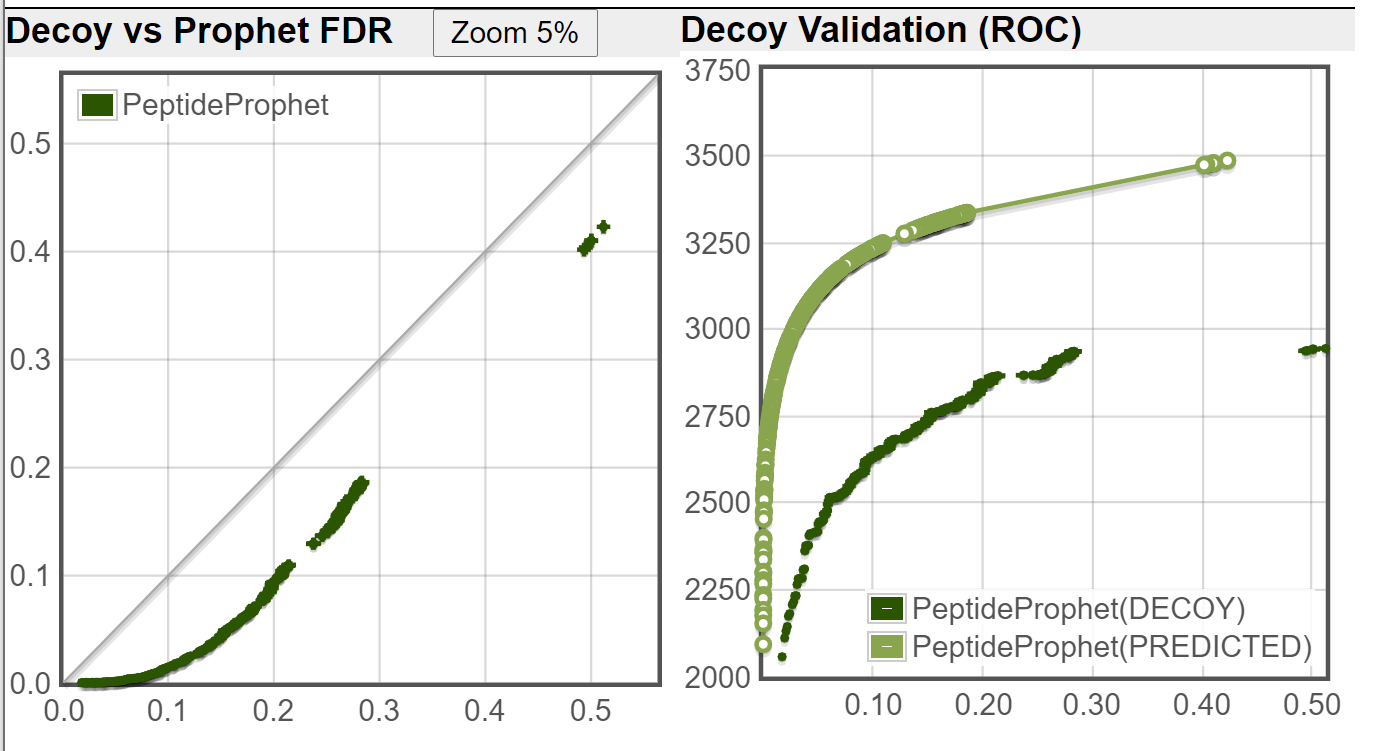
I ran the Decoy Peptide Validation tool on both of the file you sent, using DECOY1 as decoy (ignoring DECOY0 that was used by PeptideProphet), the decoy rate is applied is 1/3 (DECOY1 are about 1/3 of the fasta database when DECOY0 are ignored.) These are the results I got:
New TPP:

Old TPP:

As you can see from your decoy counts and from the above images, the new TPP is doing a much better job of controlling the error rate (as compared to an independent decoy set) on this dataset.
If you want to generate these images yourself you can use the tool in Petunia menu under "TPP Tools"->"Decoy Tools"->"Decoy Peptide Validation"
Let me know if you have any questions.
Cheers,
-David
To view this discussion on the web visit https://groups.google.com/d/msgid/spctools-discuss/60676d7d-491f-46a4-b3f6-80f5d4a1c100n%40googlegroups.com.
Reply all
Reply to author
Forward
0 new messages
