PyMedPhys Activity Report -- The new PyMedPhys GUI
PyMedPhys
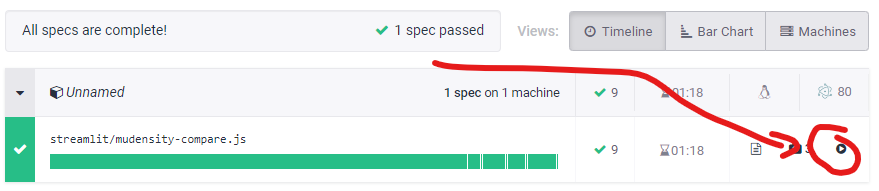
Of note, in order to roll out this GUI within our centre we have implemented automated GUI testing with Cypress (https://www.cypress.io/). This means that for every code change undergone within the PyMedPhys repo a machine spins up and runs the GUI to make sure that key tasks within the GUI have not been broken by those code changes. For example, for the 0.25.1 release, you can watch a video online of those automated tests running by going to https://dashboard.cypress.io/projects/tgt8f6/runs/57/specs and then pressing the video play button as in the screenshot below:
- 0.22.0 -- https://github.com/pymedphys/pymedphys/releases/tag/v0.22.0
- 0.23.0 -- https://github.com/pymedphys/pymedphys/releases/tag/v0.23.0
- 0.24.0 -- https://github.com/pymedphys/pymedphys/releases/tag/v0.24.0
- 0.24.1 -- https://github.com/pymedphys/pymedphys/releases/tag/v0.24.1
- 0.24.2 -- https://github.com/pymedphys/pymedphys/releases/tag/v0.24.2
- 0.24.3 -- https://github.com/pymedphys/pymedphys/releases/tag/v0.24.3
- 0.25.0 -- https://github.com/pymedphys/pymedphys/releases/tag/v0.25.0
- 0.25.1 -- https://github.com/pymedphys/pymedphys/releases/tag/v0.25.1
LipTeck Chew
1. I am confused with iComLogFiles directory and the iComLogFiles\live directory described
May I know what the differences are?
What are the recommendation for the location of logfiles, live and grouped patient directory: local or NAS?
2. In the example of listeners at https://nbviewer.jupyter.org/url/simonbiggs.net/icom.ipynb, both batch scripts consolidate the logfiles from two linacs' NSS to D:\iComLogFilesThen, in the patient archiving (grouping) it does something using D:\iComLogFiles\live and Z:\iCom\live to produce something at D:\iComLogFiles\patients
May I know what is “Z:\iCom\live” ?
Is Z:\ a local drive or network drive?
Can a single computer listen to both NSS simultaneously or we need to deploy one listener per computer for each NSS?
3. In the process of grouping patients at https://nbviewer.jupyter.org/url/simonbiggs.net/icom.ipynb, there is a line: “This is done by setting up a .bat file to run on machine boot”.
Could you elaborate further?
Is the machine here referring to the linacs or physics-server/pc?
Do you mean patient archiving (grouping) is done daily in the morning?
3. Regarding the command line tool at https://docs.pymedphys.com/ref/cli/logfile.html: pymedphys logfile orchestration [-h] [-m MOSAIQ_SQL] [-l LINAC_DETAILS] data_directory (orchestration.bat)
May I know whether this data_directory referring to ~.\pymedphys\data or D:\iComLogFiles\patients or the TRF directory?
Which data directory should we configure?
Do we need to map network drive to each separate NSS with the rtduser?
4. How do we enable reading all clinics or designated clinics appear under the \\FocalData\Installation directory described at https://github.com/pymedphys/pymedphys/blob/master/examples/site-specific/cancer-care-associates/config.toml? We have several folders for different groups of doctors.
5. Do we need the D:\iComLogFiles\patients for the gui to read trf ? What are the configurations required just to make the trf portion work? Would the trf files from two NSS override each other?
Thank you for your patience :)
warm regards,
LipTeck
Simon Biggs
Hi LipTeck,
I’m keen to send you some nice responses to this. Currently quite under the pump, might have to respond to you tonight when I get home from work.
Cheers,
Simon
--
You received this message because you are subscribed to the Google Groups "PyMedPhys" group.
To unsubscribe from this group and stop receiving emails from it, send an email to
pymedphys+...@googlegroups.com.
To view this discussion on the web, visit
https://groups.google.com/d/msgid/pymedphys/c038999e-5dc5-479b-aa58-4355ed687f27%40googlegroups.com.
Simon Biggs
Hi LipTeck,
As a first point, the iCOM command line tool recently underwent a small revamp. See https://github.com/pymedphys/pymedphys/releases/tag/v0.28.0 for more details. The GUI itself has undergone a change too, see https://github.com/pymedphys/pymedphys/releases/tag/v0.29.0.
As such, I would recommend updating to pymedphys==0.29.1 before implementing the feedback below.
Also, given this is going down the path of implementing the iCOM listener please be aware of the following bug that appears to plague all iCOM listener tools (commercial tools included) https://github.com/pymedphys/pymedphys/issues/849.
Cheers,
Simon
Hi Simon,
I have installed pymedphys and ImageMagick both in 64 bits Windows 7 and 64 bits Ubuntu 20.04 LTS Desktop versions running good in demo mode. I am now trying to setup in the actual clinical environment. Our department is using full Elekta Solution: Mosaiq v2.64, Monaco v5.11.03 with 2 Agility Head Linacs: 1 VersaHD and 1 Infinity.
1. I am confused with iComLogFiles directory and the iComLogFiles\live directory described
May I know what the differences are?
Both the live and the patients directories within the iCOM directory are for storing iCOM data. The live directory is a simple .txt dump of the most recent iCOM stream records. These files a regularly overwritten. The patients directory is where iCOM stream data is grouped and indexed by patient.
What are the recommendation for the location of logfiles, live and grouped patient directory: local or NAS?
I would have the iCOM files be stored on the same machine that is running the iCOM listener, and then have these files shared via the network so that they are accessible by the machine that is running pymedphys gui.
2. In the example of listeners at https://nbviewer.jupyter.org/url/simonbiggs.net/icom.ipynb, both batch scripts consolidate the logfiles from two linacs' NSS to D:\iComLogFilesThen, in the patient archiving (grouping) it does something using D:\iComLogFiles\live and Z:\iCom\live to produce something at D:\iComLogFiles\patients
May I know what is “Z:\iCom\live” ?
Is Z:\ a local drive or network drive?
Z:\ is a network drive shared via intranet from another site. We run this tool for 4 Linacs at 3 sites.
Can a single computer listen to both NSS simultaneously or we need to deploy one listener per computer for each NSS?
A single computer can listen to both NSS’. We have three computers doing it since there is one computer at each site and I don’t want the internet connection to cause the dropout issues discussed above.
3. In the process of grouping patients at https://nbviewer.jupyter.org/url/simonbiggs.net/icom.ipynb, there is a line: “This is done by setting up a .bat file to run on machine boot”.
Could you elaborate further?
Is the machine here referring to the linacs or physics-server/pc?
Do you mean patient archiving (grouping) is done daily in the morning?
Actually, if you install pymedphys 0.29.1, this archiving tool is no longer needed (and the CLI interface has been removed). It is now undergone automatically by the listener process already. No need to manage any of that part yourself any longer.
3. Regarding the command line tool at https://docs.pymedphys.com/ref/cli/logfile.html: pymedphys logfile orchestration [-h] [-m MOSAIQ_SQL] [-l LINAC_DETAILS] data_directory (orchestration.bat)
May I know whether this data_directory referring to ~.\pymedphys\data or D:\iComLogFiles\patients or the TRF directory?
That data directory is actually where ever you would like it to be... I personally put it on the D drive of a server’s local machine where that D drive points to a rather large spinning disk storage. The TRF files end up taking quite a lot of space.
Which data directory should we configure?
In your config.toml point to the same directory as written within the logfile orchestration tool.
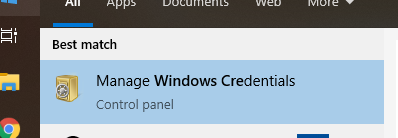
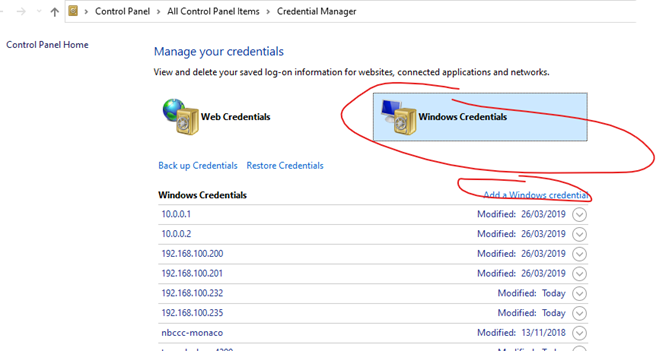
Do we need to map network drive to each separate NSS with the rtduser?
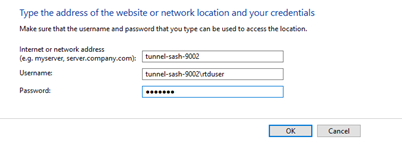
No, don’t map the NSS network drive, but do save the rtduser password within the Windows credential manager:
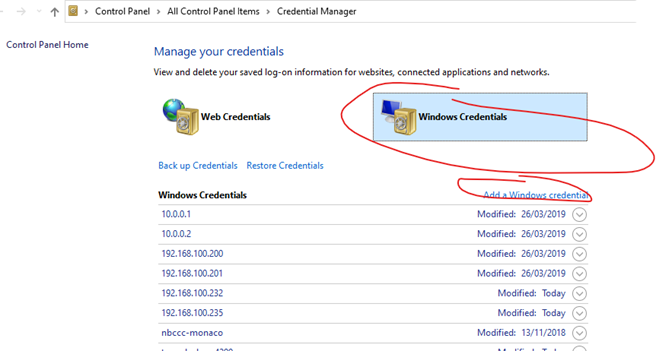
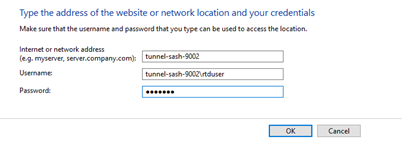
Instead of writing “tunnel-sash-9002”, instead write in the IP address of the NSS on both the network address section, and the username section.
4. How do we enable reading all clinics or designated clinics appear under the \\FocalData\Installation directory described at https://github.com/pymedphys/pymedphys/blob/master/examples/site-specific/cancer-care-associates/config.toml? We have several folders for different groups of doctors.
Unfortunately this is not currently directly supported at the moment, I might be able to make this possible. I do wonder if it might “just work” if you treat each clinic as its own “site” within the config file, as in create a new site for each clinic, and have all the configuration be the same except change the clinic path. It would be nice to directly support this use case though.
5. Do we need the D:\iComLogFiles\patients for the gui to read trf ? What are the configurations required just to make the trf portion work?
Nope, you don’t need that patient directory if you’re going to be just using TRF. However, now that patient indexing is included within the listener maybe this is not as much of an issue?
Would the trf files from two NSS override each other?
They shouldn’t. The TRF files are organised by IP address. Let me know if you have any issues here though.
Thank you for your patience :)
My pleasure :) Things are quite hectic at the moment, thanks for baring with response delays :). Feel free to let me know if you have any more issues.
warm regards,
LipTeck
LipTeck Chew
Simon Biggs
--
You received this message because you are subscribed to the Google Groups "PyMedPhys" group.
To unsubscribe from this group and stop receiving emails from it, send an email to pymedphys+...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/35511b23-3edf-4765-a3ec-58fa81ce719d%40googlegroups.com.
LipTeck CHEW
Simon Biggs
--
You received this message because you are subscribed to the Google Groups "PyMedPhys" group.
To unsubscribe from this group and stop receiving emails from it, send an email to pymedphys+...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkPH5k7vm-AtrKrPXRJh6YajBCaZAis6E-vKHMxK69h89Q%40mail.gmail.com.
LipTeck CHEW
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CAALqB0BWYiAV0nRYjSfBD6eui-5Ww_CJVHD_XxWMDq4btdwBow%40mail.gmail.com.
Simon Biggs
--
You received this message because you are subscribed to the Google Groups "PyMedPhys" group.
To unsubscribe from this group and stop receiving emails from it, send an email to pymedphys+...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkPx0ms9KBNkR_mE6hPamsNbJD77m49zrh5o_dqDefUuSQ%40mail.gmail.com.
LipTeck CHEW
Simon Biggs
Hi SimonWe have just upgraded NSS from v3 to v4 on 10-May-2020. Hope it is not NSS version dependent.regards,LipTeckOn Mon, 1 Jun 2020 at 14:29, LipTeck CHEW <liptec...@gmail.com> wrote:Hi SimonPlease see the attached text file.regards,LipTeck
--
You received this message because you are subscribed to the Google Groups "PyMedPhys" group.
To unsubscribe from this group and stop receiving emails from it, send an email to pymedphys+...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkODbrKcGoi3PE1Ur2nmX6NdPHSQ3yOGMbFSAKb2edKFuA%40mail.gmail.com.
LipTeck CHEW
Simon Biggs
Hi LipTeck,
So that bug never sees the light of day again would I be able to get a bit of data from you? Should it be fixed, would you be able to run a warm up beam or service mode beam (one that doesn’t have any patient data in it) and then, within the patients directory find the corresponding .xz file (under that particular dummy patient name). And I would be keen for you to upload this file for use within PyMedPhys’ automated testing suite. This will make sure any iCOM features that are particular to that particular file will always be verified as being able to be decoded with each code submission to PyMedPhys.
In preparation for uploading I’d like you to take a scan of the contents (so that you are happy with what you are uploading). To read the file, you will need to first extract it.
If you run “ipython” in the directory where the .xz file exists then you can run the following code within ipython:
In [1]:
import
lzma
In [2]:
import
pathlib
In [3]:
icom_paths = pathlib.Path('.').glob('*.xz')
In [4]:
for icom_path
in icom_paths:
...:
with lzma.open(icom_path,
"r")
as f:
...:
contents = f.read()
...:
...: with
open(icom_path.with_suffix(".txt"),
"wb")
as f:
...:
f.write(contents)
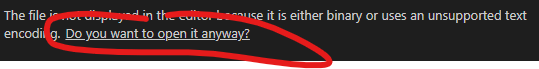
This will then convert all of the xz files to .txt files. You can then open the .txt files with something like VSCode (https://code.visualstudio.com/). When you open the text file with VSCode you will need to press “Do you want to open it anyway?”:
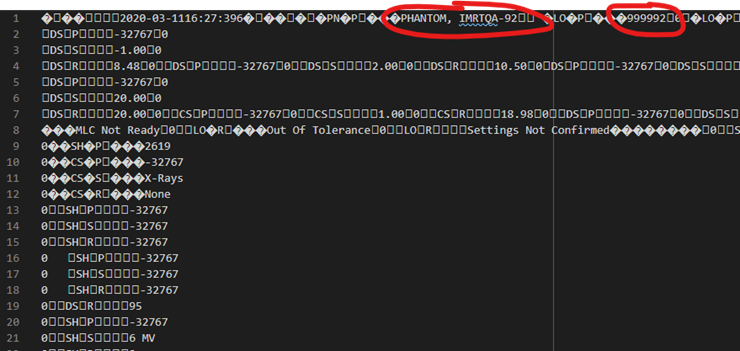
There will be lots of “boxes” and weird characters, but all patient identification information will appear in sections that look like the following:
Once you’re confident that the xz file doesn’t have any patient information within it would you then be able to use the following link to upload the .xz file to the Zenodo repository:
https://zenodo.org/deposit/new?c=pymedphys
If you have issues with Zenodo, feel free to email that .xz file on this mailing list and I will upload it.
Thanks LipTeck,
Cheers,
Simon
From: pyme...@googlegroups.com [mailto:pyme...@googlegroups.com]
On Behalf Of LipTeck CHEW
Sent: Monday, 1 June 2020 8:08 PM
To: Simon Biggs
Cc: PyMedPhys
Subject: Re: PyMedPhys Activity Report -- The new PyMedPhys GUI
Hi Simon
--
You received this message because you are subscribed to the Google Groups "PyMedPhys" group.
To unsubscribe from this group and stop receiving emails from it, send an email to
pymedphys+...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkNmLBMu4vgqAZB60HCHps0KEq_Vwbyfu0NHf-vho_bmZw%40mail.gmail.com.
LipTeck CHEW
Simon Biggs
--
You received this message because you are subscribed to the Google Groups "PyMedPhys" group.
To unsubscribe from this group and stop receiving emails from it, send an email to pymedphys+...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkMxcVypmC8UXUbYp7i-4oKiups32tXr3xdsd8w3ws3_CA%40mail.gmail.com.
LipTeck CHEW
Simon Biggs
--
You received this message because you are subscribed to the Google Groups "PyMedPhys" group.
To unsubscribe from this group and stop receiving emails from it, send an email to pymedphys+...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkNsztG2%2B0vWT16AKZs6wCNX2BNcgB6BLh2K9Y4Txx3X5w%40mail.gmail.com.
Simon Biggs
LipTeck CHEW
From: LipTeck CHEW <liptec...@gmail.com>
Date: Tue, 2 Jun 2020 at 14:36
Subject: Re: PyMedPhys Activity Report -- The new PyMedPhys GUI
Simon Biggs
--
You received this message because you are subscribed to the Google Groups "PyMedPhys" group.
To unsubscribe from this group and stop receiving emails from it, send an email to pymedphys+...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkOJ24b7YPHqQdvd%3Dj1x4P_A62LR7vjq3hDBe4O0u_nPeg%40mail.gmail.com.
Simon Biggs
Hi LipTeck,
I can reproduce your error with the file you provided by running the following:
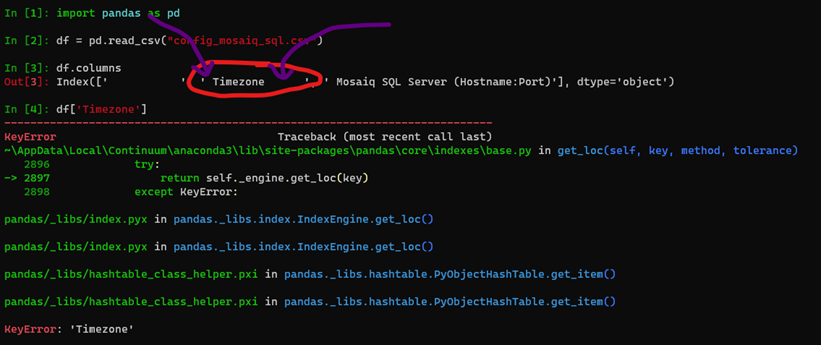
It appears the issue is due to the spaces within the csv file.
I have reattached the config file you provided just with those spaces removed. Let me know if that fixes your issues.
![]()
-->
![]()
I’ll need to update the docs (https://docs.pymedphys.com/ref/cli/logfile.html#config-mosaiq-sql-csv). It seems my best intentions to make the displayed csv files be human readable resulted in an issue. I suspect I might go down the path of using the `.pymedphys/config.toml` file for this configuration also. I think that might be a more robust and less segmented configuration choice.
Make sure to keep an eye on the release notes (https://docs.pymedphys.com/release-notes.html) whenever you upgrade just to be abreast of what parts of your set up might be adjusted. Because the version number is still 0.*.* PyMedPhys is still going to be going through quite a few adjustments before its configuration API (and other APIs) stabilise.
Cheers,
Simon
LipTeck CHEW
Simon Biggs
Hi LipTeck,
Have a look at the following file:
There, within the BAT file, I am adding python and pymedphys to the path before calling pymedphys.
Cheers,
Simon
From: LipTeck CHEW [mailto:liptec...@gmail.com]
Sent: Wednesday, 3 June 2020 10:54 AM
To: Simon Biggs
Cc: Simon Biggs; PyMedPhys
Subject: Re: PyMedPhys Activity Report -- The new PyMedPhys GUI
Hi Simon
LipTeck CHEW
Simon Biggs
Hi LipTeck,
Could you install the following development release and try again:
pip install pymedphys==0.30.0dev6
In your robo copy, you can just dump trf files anywhere within “to_be_indexed” it’ll deal with that. No need to split them up by linac.
Cheers,
Simon
From: LipTeck CHEW [mailto:liptec...@gmail.com]
Sent: Wednesday, 3 June 2020 11:11 AM
To: Simon Biggs
Cc: PyMedPhys
Subject: Re: PyMedPhys Activity Report -- The new PyMedPhys GUI
Hi Simon
LipTeck CHEW
LipTeck CHEW
LipTeck CHEW
File "c:\users\physics\miniconda3\lib\site-packages\pymedphys\_utilities\transforms\bipolar.py", line 43, in convert_IEC_angle_to_bipolar
), "Unable to automatically determine whether angle is 180 or -180"
Simon Biggs
--
You received this message because you are subscribed to the Google Groups "PyMedPhys" group.
To unsubscribe from this group and stop receiving emails from it, send an email to pymedphys+...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkMXj9_mVG75m%2BZsD4%3DnW1-a3F4ZctLxM7WKDgHwTvsrBw%40mail.gmail.com.
LipTeck CHEW
Simon Biggs
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkMWLxRT3p0TFY77sGdTbvcUF3zHWA1UB5-C-Srm4c5GPQ%40mail.gmail.com.
LipTeck CHEW
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CAALqB0Djd%3DdFuoS3uvX7TtWL8VYqBSFefsCmxQV10zo7xeArtA%40mail.gmail.com.
Simon Biggs
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkNaJjmK8TNO8ypNOHZgA79hJb_fT%2BQwy7yZVhH_MztFVQ%40mail.gmail.com.
LipTeck CHEW
Simon Biggs
That’s interesting. Are you able to deliver with Mosaiq your morning warm up beams or something like that? Or deliver a beam with a dummy patient, and then provide me the debug logs during that delivery?
By the way, in order for the file to be archived the beam needs to finish, and then the beam needs to be exited out of within Mosaiq.
From: LipTeck CHEW [mailto:liptec...@gmail.com]
Sent: Thursday, 4 June 2020 1:38 PM
To: Simon Biggs
Cc: PyMedPhys; Simon Biggs
Subject: Re: PyMedPhys Activity Report -- The new PyMedPhys GUI
Hi Simon
LipTeck CHEW
Simon Biggs
LipTeck CHEW
All good, probably can't help just now. I'll let you send through the logs on the mailing list, and see how we go. It does feel like we're getting there. Lots of hurdles though. Thanks for hanging in there :).On Thu., 4 Jun. 2020, 6:31 pm LipTeck CHEW, <liptec...@gmail.com> wrote:Sorry, just saw this. Yes, I am available.On Thu, 4 Jun 2020 at 14:57, Simon Biggs <m...@simonbiggs.net> wrote:Actually can we just quickly sort this out on a video call? Could you jump on https://whereby.com/pymedphys ?
Simon Biggs
Sent: 04 June 2020 18:52
To: Simon Biggs <m...@simonbiggs.net>; PyMedPhys <pyme...@googlegroups.com>
LipTeck CHEW
Simon Biggs
Hi LipTeck,
I have created a new dev version where the only changes that affect icom listening is that I have added a bucket more debug logging messages in and around the patient archiving logic. If you could install the new dev version and then provide me the CLI output logs that encompass two different morning run up beams (ones without patient info).
Install new version with:
pip install pymedphys==0.30.0dev7
Then run your icom listen in debug mode. That would be really helpful.
We’re getting there :).
Thanks LipTeck :),
Simon
LipTeck CHEW
Simon Biggs
Did you run it in debug mode with the latest version?
LipTeck CHEW
Simon Biggs
Ahh, can you not run –verbose, and just run –debug.
Simon Biggs
Were you actually beaming on during those logs?
Simon Biggs
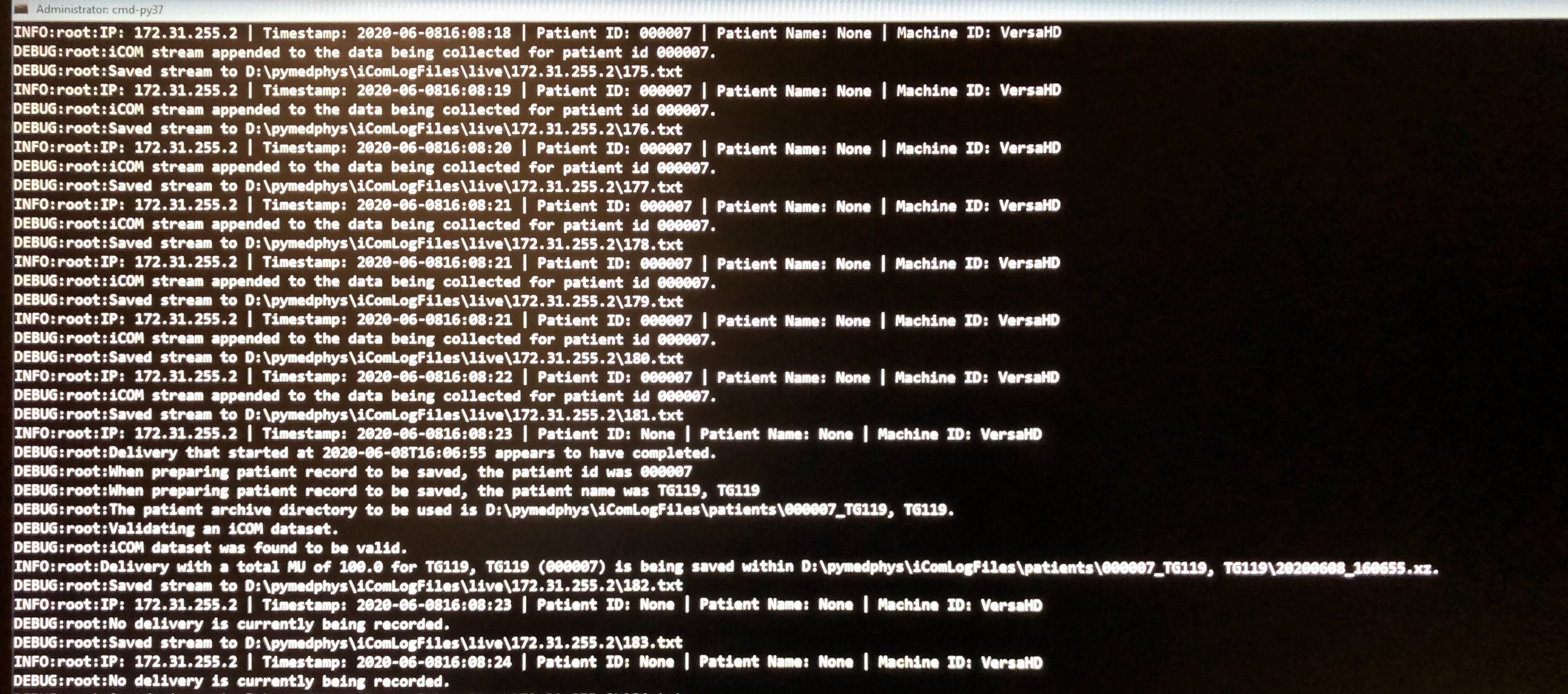
Actually, I see that you were beaming on as there are multiple iCOM streams per second, that, in my experience, only occurs during beam on time. The issue appears to be it’s not picking up the Patient ID. When you get a line like the following:
DEBUG:root:Saved stream to D:\pymedphys\iComLogFiles\live\172.31.255.2\054.txt
INFO:root:IP: 172.31.255.2 | Timestamp: 2020-06-0510:11:38 | Patient ID: None | Patient Name: Morning QA 2018, Versa | Machine ID: VersaHD
Can you stop the logging, and send me that .txt file? I would like to be able to upload that stream to be an item tested within PyMedPhys. I need to get to the bottom of why it can’t read the Patient ID within that file.
Cheers,
Simon
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/SYCP282MB05412FE99DD3692ED39D9D4095860%40SYCP282MB0541.AUSP282.PROD.OUTLOOK.COM.
Simon Biggs
Oh wow, okay. Can you try deliver a beam that does have an ID? I may need to see if there is a way I can handle “no-id” beams. Currently the software will not be able to handle a beam that does not have an ID defined. (It groups beams by Patient ID...).
From: LipTeck CHEW [mailto:liptec...@gmail.com]
Sent: Friday, 5 June 2020 1:45 PM
To: Simon Biggs
Subject: Re: PyMedPhys Activity Report -- The new PyMedPhys GUI
I have just realised there was no id. Please see screenshot attached.
However, our patients has FP numbers with prefix FP and 8 digits.
e.g.
FP12345678
FP00005678
FP12340000
On Fri, 5 Jun 2020 at 11:25, Simon Biggs <sbi...@riverinacancercare.com.au> wrote:
What is the actual patient id for that plan?
From: LipTeck CHEW [mailto:liptec...@gmail.com]
Sent: Friday, 5 June 2020 1:19 PM
To: Simon Biggs
Subject: Re: PyMedPhys Activity Report -- The new PyMedPhys GUI
Hi Simon
054.txt attached.
regards,
LipTeck
On Fri, 5 Jun 2020 at 11:04, Simon Biggs <sbi...@riverinacancercare.com.au> wrote:
All good, I’m not after the CLI log, actually importantly, the .txt file that is saved within the live directory:
D:\pymedphys\iComLogFiles\live\172.31.255.2
From: LipTeck CHEW [mailto:liptec...@gmail.com]
Sent: Friday, 5 June 2020 1:02 PM
To: Simon Biggs
Subject: Re: PyMedPhys Activity Report -- The new PyMedPhys GUI
Hi Simon,
Yes, We've loaded the morning QA patient, beam on, a 6x followed by a 10x. The lines went really fast.
I saw the format in the format below. Omitting the lines on the 000-255 files:
Mosaiq beam loaded:
| Patient Name: Morning QA 2018, Versa | Machine ID: VersaHD
| Patient Name: Morning QA 2018, Versa | Machine ID: VersaHD
| Patient Name: Morning QA 2018, Versa | Machine ID: VersaHD
Beam on:
| Patient Name: None | Machine ID: VersaHD
| Patient Name: None | Machine ID: VersaHD
| Patient Name: Morning QA 2018, Versa | Machine ID: VersaHD
| Patient Name: Morning QA 2018, Versa | Machine ID: VersaHD
during beam on:
| Patient Name: None | Machine ID: VersaHD
| Patient Name: None | Machine ID: VersaHD
| Patient Name: None | Machine ID: VersaHD
| Patient Name: None | Machine ID: VersaHD
beam off:
| Patient Name: Morning QA 2018, Versa | Machine ID: VersaHD
| Patient Name: Morning QA 2018, Versa | Machine ID: VersaHD
| Patient Name: None | Machine ID: VersaHD
| Patient Name: None | Machine ID: VersaHD
it's too fast to catch the complete log
regards,
LipTeck
LipTeck CHEW
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/SYCP282MB05414D85600D2D9C67B252B295860%40SYCP282MB0541.AUSP282.PROD.OUTLOOK.COM.
LipTeck CHEW
Simon Biggs
Unfortunately I can only make use of what the iCOM stream provides. If you open up those text files with VSCode you can search for whether or not any of those IDs appear within that file.
LipTeck CHEW
On Sat, 6 Jun 2020 at 11:17 AM, LipTeck CHEW <liptec...@gmail.com> wrote:
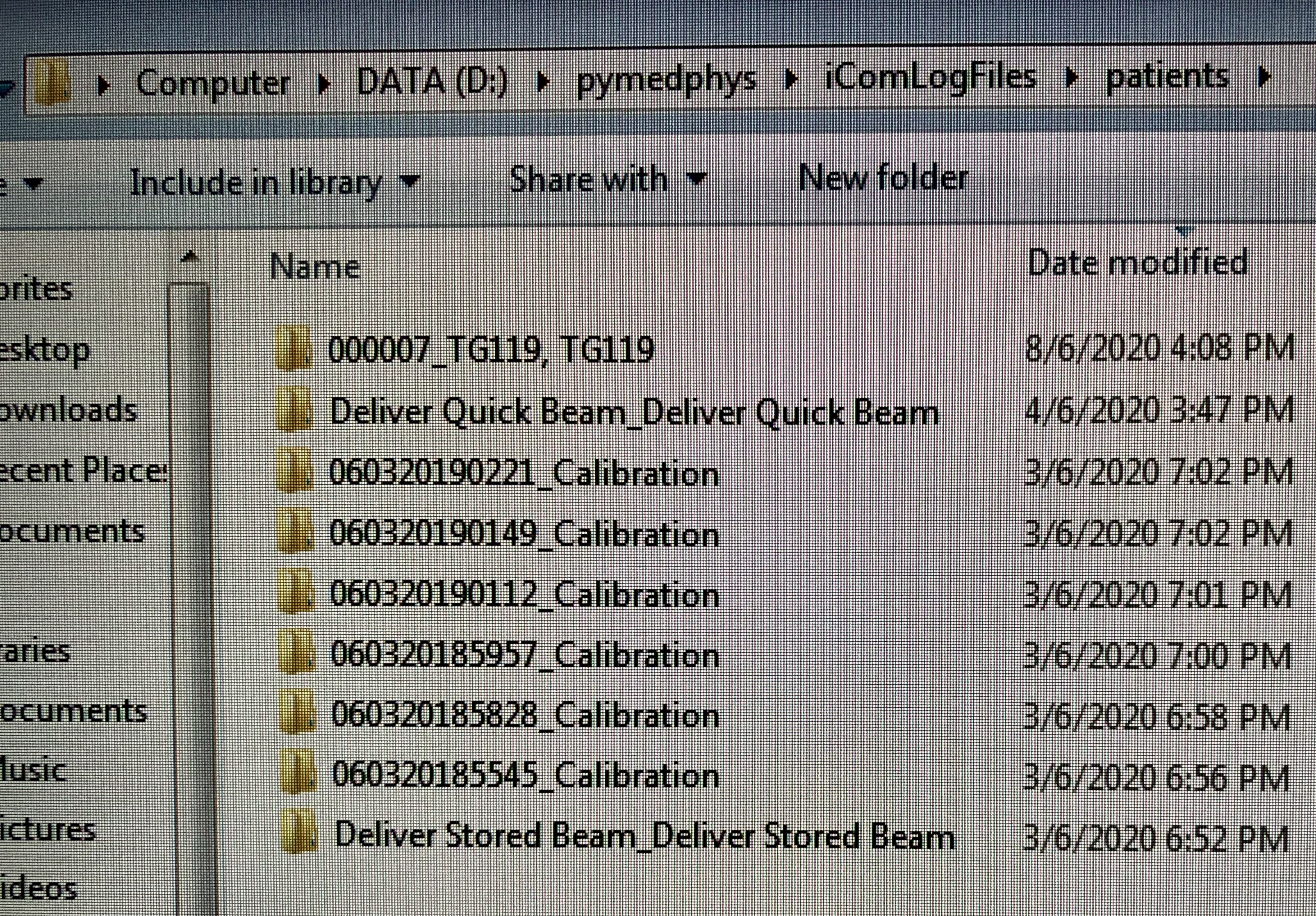
Hi Simonre: pymedphys icom listenSorry that we were occupied with a few new cases.I share with you one of the Mobius elog file in year 2019 for a pelvis vmat case. I have edited the PatientName, PatientID, and TreatmentName. I have excluded the ebin file that has the actual patient information.The elog file name begins with time stamp followed by patient Sirname and then separated by comma without space with first name and then one space with FP number in bracket. It is able to read the FP as the PatientID. Therefore, I think this information is available in the iCom.It is interesting that the Mobius log use the time stamp as the start of the filename and then adds a bracket to the PatientID to end the filename:"2019-07-03, 11-18-45, Sirname,Firstname (FP12345678).txt"Perhaps in this way, it is able to handle blank fields. So, a Morning QA 2018, Versa patient without FP would look like this:"yyyy-mm-dd, tt-hh-ss, Morning QA 2018,Versa ().txt"Generally Mobius logs everything but only logs that have PatientID, FieldID, and TreatmenName are useful for the MobiusFX. Perhaps combining PatientID, FieldID and TreatmentName together with something to create a ReferenceID for regrouping will make the icom listener much more robust.re: pymedphys orchestrationThe new user account for Mosaiq db_reader will be provided on next Tueday 9-June-2020.have a good weekend!regards,LipTeck
Simon Biggs
Brilliant, that’s exactly what I was hoping to see :).
Cheers,
LipTeck CHEW
Simon Biggs
--
You received this message because you are subscribed to the Google Groups "PyMedPhys" group.
To unsubscribe from this group and stop receiving emails from it, send an email to pymedphys+...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkPbsKVFokEc3npnbUwLGqO4iFYEQJrAQj9DDTTrrGOHDg%40mail.gmail.com.
Simon Biggs
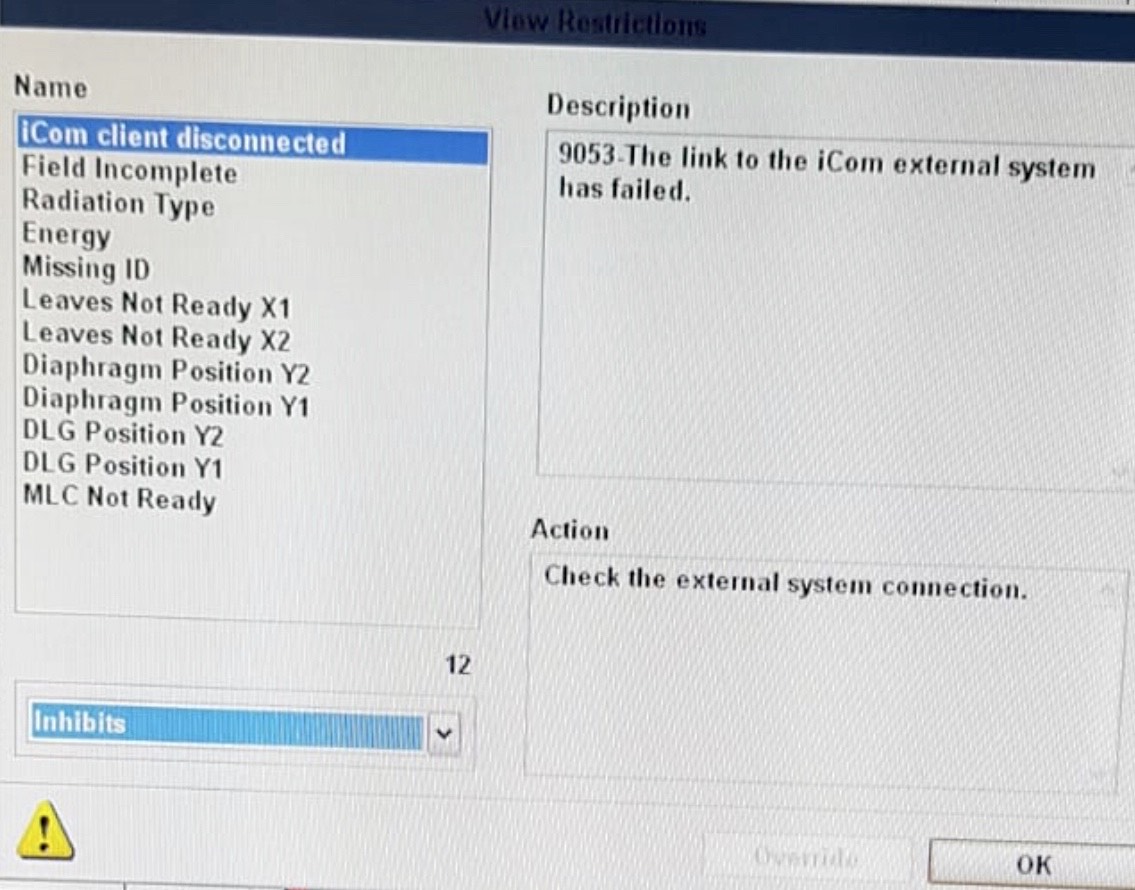
I think the icom listen CLI has been reading the FP number and necessary information but what are the other criteria that were not met causing it not to group? None of the patients were grouped. The difference of the actual patient treatment with the above TG119, TG119 and Deliver Quick Beam_Deliver Quick Beam is patient setup imaging and the total time of all possible beams under one prescription. Is there a limit of time between fields or total time for each prescription?
Regarding the 000-255 files. Could you explain how the data is logged? Since the information is coming at 0.25 sec, how is the data recorded in these 256 files? What happens after 256 or 64 sec?
LipTeck CHEW
Simon Biggs
My pleasure :) I do have a favour to ask. Once this is all over, would you be interested in collating your installation notes into a document? I can then upload that document to help guide others through what you’ve achieved :).
Let me know if you’re interested.
Cheers,
Simon
From: LipTeck CHEW [mailto:liptec...@gmail.com]
Sent: Wednesday, 10 June 2020 9:24 AM
To: Simon Biggs
Cc: PyMedPhys; Simon Biggs
Subject: Re: PyMedPhys Activity Report -- The new PyMedPhys GUI
Hi Simon
LipTeck CHEW
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/SYCP282MB0541A6E67CEBA4D5D558C74795820%40SYCP282MB0541.AUSP282.PROD.OUTLOOK.COM.
Simon Biggs
That would be quite cool. In the past I have thought it would be quite neat to put together pre-built raspberry pi’s for people, maybe have a little “PyMedPhys” shop (it could sell shirts too :) ). If you like that idea, it might be interesting setting something up together.
LipTeck CHEW
LipTeck CHEW
Simon Biggs
Are you able to make a dummy patient that has the same ID format as your clinical patients and reproduce the effect in logs that you are able to share?
From: LipTeck CHEW [mailto:liptec...@gmail.com]
Sent: Wednesday, 10 June 2020 11:16 AM
To: Simon Biggs
Cc: PyMedPhys; Simon Biggs
Subject: Re: PyMedPhys Activity Report -- The new PyMedPhys GUI
Hi Simon
LipTeck CHEW
LipTeck CHEW
LipTeck CHEW
Simon Biggs
--
You received this message because you are subscribed to the Google Groups "PyMedPhys" group.
To unsubscribe from this group and stop receiving emails from it, send an email to pymedphys+...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkOaOGEnUeA_7vcyGqX2jb62JVuAnA4RVjHCZ%3DOCOvxXHg%40mail.gmail.com.
LipTeck CHEW
Simon Biggs
--
You received this message because you are subscribed to the Google Groups "PyMedPhys" group.
To unsubscribe from this group and stop receiving emails from it, send an email to pymedphys+...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkP9KdTpYPrR%3DdnVURzMiTWN5Q-7sL%2BtL66gS%2BK9Jh8R8g%40mail.gmail.com.
LipTeck CHEW
Hi LipTeck,
As a first point, the iCOM command line tool recently underwent a small revamp. See https://github.com/pymedphys/pymedphys/releases/tag/v0.28.0 for more details. The GUI itself has undergone a change too, see https://github.com/pymedphys/pymedphys/releases/tag/v0.29.0.
As such, I would recommend updating to pymedphys==0.29.1 before implementing the feedback below.
Also, given this is going down the path of implementing the iCOM listener please be aware of the following bug that appears to plague all iCOM listener tools (commercial tools included) https://github.com/pymedphys/pymedphys/issues/849.
Cheers,
Simon
Hi Simon,
I have installed pymedphys and ImageMagick both in 64 bits Windows 7 and 64 bits Ubuntu 20.04 LTS Desktop versions running good in demo mode. I am now trying to setup in the actual clinical environment. Our department is using full Elekta Solution: Mosaiq v2.64, Monaco v5.11.03 with 2 Agility Head Linacs: 1 VersaHD and 1 Infinity.
1. I am confused with iComLogFiles directory and the iComLogFiles\live directory described
May I know what the differences are?
Both the live and the patients directories within the iCOM directory are for storing iCOM data. The live directory is a simple .txt dump of the most recent iCOM stream records. These files a regularly overwritten. The patients directory is where iCOM stream data is grouped and indexed by patient.
What are the recommendation for the location of logfiles, live and grouped patient directory: local or NAS?
I would have the iCOM files be stored on the same machine that is running the iCOM listener, and then have these files shared via the network so that they are accessible by the machine that is running pymedphys gui.
2. In the example of listeners at https://nbviewer.jupyter.org/url/simonbiggs.net/icom.ipynb, both batch scripts consolidate the logfiles from two linacs' NSS to D:\iComLogFilesThen, in the patient archiving (grouping) it does something using D:\iComLogFiles\live and Z:\iCom\live to produce something at D:\iComLogFiles\patients
May I know what is “Z:\iCom\live” ?
Is Z:\ a local drive or network drive?
Z:\ is a network drive shared via intranet from another site. We run this tool for 4 Linacs at 3 sites.
Can a single computer listen to both NSS simultaneously or we need to deploy one listener per computer for each NSS?
A single computer can listen to both NSS’. We have three computers doing it since there is one computer at each site and I don’t want the internet connection to cause the dropout issues discussed above.
3. In the process of grouping patients at https://nbviewer.jupyter.org/url/simonbiggs.net/icom.ipynb, there is a line: “This is done by setting up a .bat file to run on machine boot”.
Could you elaborate further?
Is the machine here referring to the linacs or physics-server/pc?
Do you mean patient archiving (grouping) is done daily in the morning?
Actually, if you install pymedphys 0.29.1, this archiving tool is no longer needed (and the CLI interface has been removed). It is now undergone automatically by the listener process already. No need to manage any of that part yourself any longer.
3. Regarding the command line tool at https://docs.pymedphys.com/ref/cli/logfile.html: pymedphys logfile orchestration [-h] [-m MOSAIQ_SQL] [-l LINAC_DETAILS] data_directory (orchestration.bat)
May I know whether this data_directory referring to ~.\pymedphys\data or D:\iComLogFiles\patients or the TRF directory?
That data directory is actually where ever you would like it to be... I personally put it on the D drive of a server’s local machine where that D drive points to a rather large spinning disk storage. The TRF files end up taking quite a lot of space.
Which data directory should we configure?
In your config.toml point to the same directory as written within the logfile orchestration tool.
Do we need to map network drive to each separate NSS with the rtduser?
No, don’t map the NSS network drive, but do save the rtduser password within the Windows credential manager:
Instead of writing “tunnel-sash-9002”, instead write in the IP address of the NSS on both the network address section, and the username section.
4. How do we enable reading all clinics or designated clinics appear under the \\FocalData\Installation directory described at https://github.com/pymedphys/pymedphys/blob/master/examples/site-specific/cancer-care-associates/config.toml? We have several folders for different groups of doctors.
Unfortunately this is not currently directly supported at the moment, I might be able to make this possible. I do wonder if it might “just work” if you treat each clinic as its own “site” within the config file, as in create a new site for each clinic, and have all the configuration be the same except change the clinic path. It would be nice to directly support this use case though.
5. Do we need the D:\iComLogFiles\patients for the gui to read trf ? What are the configurations required just to make the trf portion work?
Nope, you don’t need that patient directory if you’re going to be just using TRF. However, now that patient indexing is included within the listener maybe this is not as much of an issue?
Would the trf files from two NSS override each other?
They shouldn’t. The TRF files are organised by IP address. Let me know if you have any issues here though.
Thank you for your patience :)
My pleasure :) Things are quite hectic at the moment, thanks for baring with response delays :). Feel free to let me know if you have any more issues.
warm regards,
LipTeck
Simon Biggs
--
You received this message because you are subscribed to the Google Groups "PyMedPhys" group.
To unsubscribe from this group and stop receiving emails from it, send an email to pymedphys+...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkMhozOjo3T9OYov-SkaTR4%2BDL%2BeyoHyF9-XrEG_N2NGmw%40mail.gmail.com.
Simon Biggs
LipTeck CHEW
Simon Biggs
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkPPxGcoCi5ZDimzMZf0hnkcgbukx1fJPX1Dmc9OWzUi_A%40mail.gmail.com.
LipTeck CHEW
Simon Biggs
Simon Biggs
Hi LipTeck,
I imagine to be able to resolve these remaining issues in a timely fashion I would likely need to spend a consolidated block of time within work hours. Would you be open to working out a consultancy agreement with my employer so that I can be given the time + focus to fix these issues for you?
Let me know your thoughts,
Cheers,
Simon
From: pyme...@googlegroups.com [mailto:pyme...@googlegroups.com]
On Behalf Of Simon Biggs
Sent: Saturday, 13 June 2020 5:36 PM
To: PyMedPhys
Subject: Re: PyMedPhys Activity Report -- The new PyMedPhys GUI
Hi LipTeck,
--
You received this message because you are subscribed to the Google Groups "PyMedPhys" group.
To unsubscribe from this group and stop receiving emails from it, send an email to
pymedphys+...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/6363e5ca-f67d-4d4b-9a34-33d75d344efao%40googlegroups.com.
LipTeck CHEW
Simon Biggs
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkMNicV5jcy_EXpSztHv_mzqGz-Co_CkpVA%3Dx3DzpVcKgQ%40mail.gmail.com.
LipTeck CHEW
Hi LipTeck,
I can reproduce your error with the file you provided by running the following:
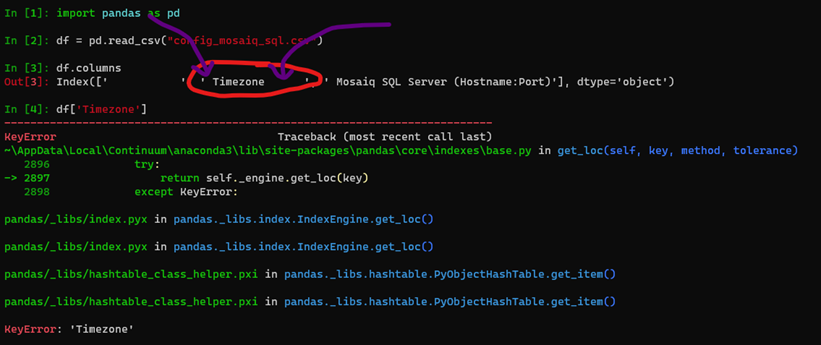
It appears the issue is due to the spaces within the csv file.
I have reattached the config file you provided just with those spaces removed. Let me know if that fixes your issues.
-->
I’ll need to update the docs (https://docs.pymedphys.com/ref/cli/logfile.html#config-mosaiq-sql-csv). It seems my best intentions to make the displayed csv files be human readable resulted in an issue. I suspect I might go down the path of using the `.pymedphys/config.toml` file for this configuration also. I think that might be a more robust and less segmented configuration choice.
Make sure to keep an eye on the release notes (https://docs.pymedphys.com/release-notes.html) whenever you upgrade just to be abreast of what parts of your set up might be adjusted. Because the version number is still 0.*.* PyMedPhys is still going to be going through quite a few adjustments before its configuration API (and other APIs) stabilise.
Cheers,
Simon
From: LipTeck CHEW [mailto:liptec...@gmail.com]
Sent: Tuesday, 2 June 2020 10:26 PM
To: Simon Biggs; PyMedPhys
Subject: Fwd: PyMedPhys Activity Report -- The new PyMedPhys GUI
Hi Simon
Is there any solution to the timezone error? Singapore is at the GMT +8. Are there alternatives to replace "Asia/Singapore" to enable the CLI to run first?
Thanks :)
regards,
LipTeck
---------- Forwarded message ---------
From: LipTeck CHEW <liptec...@gmail.com>
Date: Tue, 2 Jun 2020 at 14:36
Subject: Re: PyMedPhys Activity Report -- The new PyMedPhys GUI
To: Simon Biggs <m...@simonbiggs.net>
Hi Simon
Please see attached orchestration error text file.
regards,
LipTeck
Simon Biggs
--
You received this message because you are subscribed to the Google Groups "PyMedPhys" group.
To unsubscribe from this group and stop receiving emails from it, send an email to pymedphys+...@googlegroups.com.
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkMWrFDsn%3DFOr%2BtQOyn0gqMXy0xE6jPRUWL1sXch4E-kTg%40mail.gmail.com.
Simon Biggs
LipTeck CHEW
I find the time is incorrect. Any comments? For a successful orchestration, what do we get to see?Header(machine='3316', date='20/08/04 03:20:45 Z', timezone='+08:00', field_label='V1', field_name='Arc1')Traceback (most recent call last):File "c:\users\physics\miniconda3\lib\site-packages\pymedphys\_trf\manage\index.py", line 164, in file_ready_to_be_indexedcentre = machine_map[header.machine]["centre"]KeyError: '3316'
Simon Biggs
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkOjVTOf4NXnM7GCfyGXTKGF78r%3DyLcda%2BXNtgaqRHk6DQ%40mail.gmail.com.
LipTeck CHEW
Simon Biggs
Hi LipTeck,
To get to the bottom of the identifying issue, could you try calling the following function directly within Python:
That’s the function used to identify the TRF file internally, it takes the information from the TRF header and undergoes a Mosaiq query based on that header (and timezone offsets) in order to identify the TRF file.
LipTeck CHEW
Simon Biggs
Hi LipTeck,
To run it you want to do the following:
import pymedphys
import pymedphys._mosaiq.delivery as delivery
with pymedphys.connect(mosaiq_hostname) as cursor:
# get machine, delivery_time, field_label, field_name from the logfile header
delivery.get_mosaiq_delivery_details(cursor, machine, delivery_time, field_label, field_name, buffer=240)
Cheers,
Simon
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkPK4FPq_zUyVtBD%2BrApSP7YrnKvmhQUCD%3DY6NHTJtLyPg%40mail.gmail.com.
Simon Biggs
With pretty printing:
import pymedphys
import pymedphys._mosaiq.delivery as delivery
with pymedphys.connect(mosaiq_hostname) as cursor:
# get machine, delivery_time, field_label, field_name from the logfile header
delivery.get_mosaiq_delivery_details(cursor, machine, delivery_time, field_label, field_name, buffer=240)
From: Simon Biggs
Sent: Wednesday, 5 August 2020 3:39 PM
To: 'LipTeck CHEW'
Cc: Simon Biggs; PyMedPhys
Subject: RE: PyMedPhys Activity Report -- The new PyMedPhys GUI
Hi LipTeck,
To run it you want to do the following:
import pymedphys
import pymedphys._mosaiq.delivery as delivery
with pymedphys.connect(mosaiq_hostname) as cursor:
# get machine, delivery_time, field_label, field_name from the logfile header
delivery.get_mosaiq_delivery_details(cursor, machine, delivery_time, field_label, field_name, buffer=240)
Cheers,
Simon
To view this discussion on the web, visit https://groups.google.com/d/msgid/pymedphys/CA%2B5_vkPK4FPq_zUyVtBD%2BrApSP7YrnKvmhQUCD%3DY6NHTJtLyPg%40mail.gmail.com.
LipTeck CHEW
Simon Biggs
Hi LipTeck,
I’m not sure about spyder and what magic it is doing. Could you either try within Jupyter Lab or an ipython console prompt?
LipTeck CHEW
Simon Biggs
Yup, that’s fair.
I should have written:
pymedphys.mosaiq.connect :)