Skip to first unread message
Hollix00
Jun 11, 2019, 9:06:57 AM6/11/19
to lavaan
Hi folks,
usually I use M. Cheung's metaSEM packages to specify meta-analytical SEMs. Especially I use the wls() function. Now I would like to test equality of parameters via specifying equality constraints and found (I guess so) that this is not possible with the wls-function but only in his tssem-function. However, using tssem is not possible that my data is in a long format (multilevel structure) and not the tsssem-typical list format (in which each element is the correlation matrix of each primary study).
Long story short: I would like to incorporate the pooled correlation matrix together with the asymptotic covariance matrix (ACM) in lavaan and use the wls-estimator there. However, I have not found any clue how to do this.
Can anybody tell me how the syntax looks like (i.e., incorporating the matrices of two groups together with their ACMs) and testing for group differences?
Best,
Holger
Holger Steinmetz
Jun 11, 2019, 9:23:45 AM6/11/19
to lavaan
One follow up, after inspecting "?sem" I tried the following code, which gave me an error:
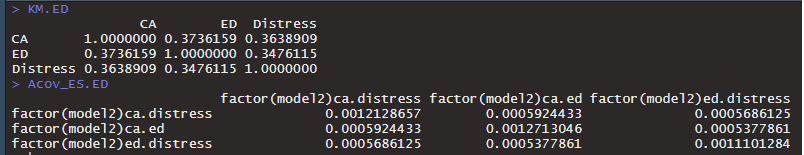
> summary(sem(mod.ed, sample.cov=KM.ED, sample.nobs = n.ED, estimator="wls", WLS.V=Acov_ES.ED))
Error in crossprod(WLS.V, diff) : non-conformable arguments
In the code (an example for one of the groups),
-- mod.ed is the SEM-structure,
-- KM.ED is a simple correlation matrix (pooled across several studies)
-- n.ED is the total N
-- Acov_ES.ED is the asymptotic covariance matrix.
Perhaps someone can point me in the right direction.
Best,
Holger
Mike Cheung
Jun 11, 2019, 9:27:49 PM6/11/19
to lav...@googlegroups.com
Hi Holger,
You may specify equality constraints by using the same parameter labels in the A and S matrices in both wls() and tssem2() in the metaSEM package.
P.S. Since the question is not related to lavaan, it may be better to post them to https://openmx.ssri.psu.edu/forums/third-party-software/metasem.
--
---------------------------------------------------------------------
Mike W.L. Cheung Phone: (65) 6516-3702
Department of Psychology Fax: (65) 6773-1843
National University of Singapore
http://mikewlcheung.github.io/
---------------------------------------------------------------------
Mike W.L. Cheung Phone: (65) 6516-3702
Department of Psychology Fax: (65) 6773-1843
National University of Singapore
http://mikewlcheung.github.io/
---------------------------------------------------------------------
--
You received this message because you are subscribed to the Google Groups "lavaan" group.
To unsubscribe from this group and stop receiving emails from it, send an email to lavaan+un...@googlegroups.com.
To post to this group, send email to lav...@googlegroups.com.
Visit this group at https://groups.google.com/group/lavaan.
To view this discussion on the web visit https://groups.google.com/d/msgid/lavaan/c7e2d2cb-395f-40ac-bcba-c39899828e8f%40googlegroups.com.
For more options, visit https://groups.google.com/d/optout.
Holger Steinmetz
Jun 12, 2019, 1:17:35 AM6/12/19
to lavaan
Dear Mike,
thank you so much for this helpful answer (and thanks also for your response on my SEMNET post. Suzanne's and your subgroup paper was the background of my question here).
With regard to your lavaan-note: I do not quite understand this note as I had a lavaan-question (how to use the wls-estimator in lavaan and I reported an error message). This is still valid.
Actually --as a heavy lavaan user-- I would like to do MASEM in lavaan :) By incorporating the ACM, this should be possible..
Best,
Holger
Am Mittwoch, 12. Juni 2019 03:27:49 UTC+2 schrieb Mike Cheung:
Hi Holger,You may specify equality constraints by using the same parameter labels in the A and S matrices in both wls() and tssem2() in the metaSEM package.P.S. Since the question is not related to lavaan, it may be better to post them to https://openmx.ssri.psu.edu/forums/third-party-software/metasem.-----------------------------------------------------------------------
Mike W.L. Cheung Phone: (65) 6516-3702
Department of Psychology Fax: (65) 6773-1843
National University of Singapore
http://mikewlcheung.github.io/
---------------------------------------------------------------------
On Tue, Jun 11, 2019 at 9:07 PM 'Hollix00' via lavaan <lav...@googlegroups.com> wrote:
Hi folks,--usually I use M. Cheung's metaSEM packages to specify meta-analytical SEMs. Especially I use the wls() function. Now I would like to test equality of parameters via specifying equality constraints and found (I guess so) that this is not possible with the wls-function but only in his tssem-function. However, using tssem is not possible that my data is in a long format (multilevel structure) and not the tsssem-typical list format (in which each element is the correlation matrix of each primary study).Long story short: I would like to incorporate the pooled correlation matrix together with the asymptotic covariance matrix (ACM) in lavaan and use the wls-estimator there. However, I have not found any clue how to do this.Can anybody tell me how the syntax looks like (i.e., incorporating the matrices of two groups together with their ACMs) and testing for group differences?Best,Holger
You received this message because you are subscribed to the Google Groups "lavaan" group.
To unsubscribe from this group and stop receiving emails from it, send an email to lav...@googlegroups.com.
Mike Cheung
Jun 12, 2019, 1:35:27 AM6/12/19
to lav...@googlegroups.com
Dear Holger,
I see. Thanks for the clarification. I thought that you were referring to the wls() function in the metaSEM package.
--
---------------------------------------------------------------------
Mike W.L. Cheung Phone: (65) 6516-3702
Department of Psychology Fax: (65) 6773-1843
National University of Singapore
http://mikewlcheung.github.io/
---------------------------------------------------------------------
Mike W.L. Cheung Phone: (65) 6516-3702
Department of Psychology Fax: (65) 6773-1843
National University of Singapore
http://mikewlcheung.github.io/
---------------------------------------------------------------------
To unsubscribe from this group and stop receiving emails from it, send an email to lavaan+un...@googlegroups.com.
To post to this group, send email to lav...@googlegroups.com.
Visit this group at https://groups.google.com/group/lavaan.
To view this discussion on the web visit https://groups.google.com/d/msgid/lavaan/35159a75-8508-4c64-b82f-30fed86a352d%40googlegroups.com.
Yves Rosseel
Jun 13, 2019, 4:44:55 AM6/13/19
to lav...@googlegroups.com
A small example below to show how use can use the wls(mv) estimator
using sample statistics only. It is a bit of work, but it can be done:
Yves.
library(lavaan)
# create binary data
HS9 <- HolzingerSwineford1939[,c("x1","x2","x3","x4","x5",
"x6","x7","x8","x9")]
Data <- as.data.frame( lapply(HS9, cut, 2, labels=FALSE) )
# simple one-factor model
model <- ' trait =~ x1 + x2 + x3 + x4 + x5 + x6 '
# single group, no covariates
fit <- sem(model, data = Data)
# data summary
sample.cov <- lavInspect(fit, "sampstat")$cov
sample.mean <- lavInspect(fit, "sampstat")$mean
sample.th <- lavInspect(fit, "sampstat")$th
attr(sample.th, "th.idx") <- lavInspect(fit, "th.idx")
sample.nobs <- lavInspect(fit, "nobs")
WLS.V <- lavInspect(fit, "WLS.V")
NACOV <- lavInspect(fit, "gamma")
# fitting same model using sample statistics only
fit.bis <- cfa(model, sample.cov = sample.cov, sample.mean =
sample.mean, sample.th = sample.th, sample.nobs = sample.nobs,
WLS.V = WLS.V, NACOV = NACOV)
fit.bis
using sample statistics only. It is a bit of work, but it can be done:
Yves.
library(lavaan)
# create binary data
HS9 <- HolzingerSwineford1939[,c("x1","x2","x3","x4","x5",
"x6","x7","x8","x9")]
Data <- as.data.frame( lapply(HS9, cut, 2, labels=FALSE) )
# simple one-factor model
model <- ' trait =~ x1 + x2 + x3 + x4 + x5 + x6 '
# single group, no covariates
fit <- sem(model, data = Data)
# data summary
sample.cov <- lavInspect(fit, "sampstat")$cov
sample.mean <- lavInspect(fit, "sampstat")$mean
sample.th <- lavInspect(fit, "sampstat")$th
attr(sample.th, "th.idx") <- lavInspect(fit, "th.idx")
sample.nobs <- lavInspect(fit, "nobs")
WLS.V <- lavInspect(fit, "WLS.V")
NACOV <- lavInspect(fit, "gamma")
# fitting same model using sample statistics only
fit.bis <- cfa(model, sample.cov = sample.cov, sample.mean =
sample.mean, sample.th = sample.th, sample.nobs = sample.nobs,
WLS.V = WLS.V, NACOV = NACOV)
fit.bis
Holger Steinmetz
Jun 13, 2019, 7:36:28 AM6/13/19
to lavaan
Hi Yves,
thanks for example. I saw that reducing your code to the sample cov, sample siize and ACM was sufficient to reproduce the individual-based CFA:
fit.bis <- cfa(model, sample.cov = sample.cov, sample.nobs = sample.nobs, WLS.V = WLS.V)
That line of code is exactly what I tried with my meta-analytical matrix (a pooled correlation matrix) and its associated ACM (that resulted from the pooling) and I get the error.
> fit.ED <- sem(mod.ed, sample.cov=KM.ED, sample.nobs = n.ED, estimator="wls", WLS.V=Acov_ES.ED)
Error in crossprod(WLS.V, diff) : non-conformable arguments
Yves Rosseel
Jun 13, 2019, 9:23:26 AM6/13/19
to lav...@googlegroups.com
The problem is with the correlation matrix: lavaan only accepts
covariance matrices (for continuous data), and when p=3, it expects ACOV
to be of dimension 6x6, but it gets 3x3.
I am afraid there is no easy fix for this one, until lavaan supports
correlations structures for continuous data.
Yves.
> Matrices.png <about:invalid#zClosurez>
> <mailto:lavaan+un...@googlegroups.com>.
> <https://groups.google.com/d/msgid/lavaan/d541e53d-a305-42ee-bb20-385521793172%40googlegroups.com?utm_medium=email&utm_source=footer>.
covariance matrices (for continuous data), and when p=3, it expects ACOV
to be of dimension 6x6, but it gets 3x3.
I am afraid there is no easy fix for this one, until lavaan supports
correlations structures for continuous data.
Yves.
>
>
>
>
> Best,
> Holger
>
> --
> You received this message because you are subscribed to the Google
> Groups "lavaan" group.
> To unsubscribe from this group and stop receiving emails from it, send
> an email to lavaan+un...@googlegroups.com
>
>
>
> Best,
> Holger
>
> --
> You received this message because you are subscribed to the Google
> Groups "lavaan" group.
> To unsubscribe from this group and stop receiving emails from it, send
> <mailto:lavaan+un...@googlegroups.com>.
> To post to this group, send email to lav...@googlegroups.com
> <mailto:lav...@googlegroups.com>.
> Visit this group at https://groups.google.com/group/lavaan.
> To view this discussion on the web visit
> https://groups.google.com/d/msgid/lavaan/d541e53d-a305-42ee-bb20-385521793172%40googlegroups.com
> To view this discussion on the web visit
> <https://groups.google.com/d/msgid/lavaan/d541e53d-a305-42ee-bb20-385521793172%40googlegroups.com?utm_medium=email&utm_source=footer>.
Terrence Jorgensen
Nov 18, 2019, 10:12:01 AM11/18/19
to lavaan
The problem is with the correlation matrix: lavaan only accepts
covariance matrices (for continuous data), and when p=3, it expects ACOV
to be of dimension 6x6, but it gets 3x3.
I am afraid there is no easy fix for this one, until lavaan supports
correlations structures for continuous data.
Holger, I figured out a hack to get around this issue, if you are still interested in performing MASEM on correlation matrices in lavaan.
The trick is to coax lavaan into expecting correlations instead of covariances among your continuous variables. This can be accomplished by lying to lavaan (shame on us!), telling it that the variables are categorical, in which case it will expect thresholds for those variables. Naturally, you will not have those, but you can get around that fact by passing an arbitrary vector of fabricated sample thresholds (which will be saturated in your model, so it won't affect the model's fit or df) and appending rows/columns to Acov_ES.ED that correspond to those (pretend) "estimated" thresholds.
Hopefully this script works for you as-is. If not, perhaps you could post your meta-data so I can work with those and provide syntax I can verify will work. FYI, my syntax below assumes Acov_ES.ED is the asymptotic sampling covariance matrix of the correlations (i.e., the SEs are the square-roots of diag(Acov_ES.ED)). If your original descriptions were correct, then Acov_ES.ED should not be the weight matrix (WLS.V). If Acov_ES.ED is actually the weight matrix (inverted NACOV), then you can start by inverting it into the NACOV to obtain what you need via the syntax below.
nVariables <- dim(KM.ED)[1]
nMoments <- dim(Acov_ES.ED)[1]
## construct NACOV, appended with 1 threshold per variable to trick lavaan
NACOV <- matrix(0, nrow = nVariables + nMoments, ncol = nVariables + nMoments)
## lavaan expects thresholds first:
## - arbitrary sampling variance = weight = 1
## - no covariance with other estimates
diag(NACOV)[1:nVariables] <- 1
## remaining elements are the estimates you have
idx <- (nVariables + 1):(nVariables + nMoments)
NACOV[idx, idx] <- n.ED * Acov_ES.ED
## diagonally WLS (since lavaan thinks these are ordered variables)
WLS.V <- MASS::ginv(diag(diag(NACOV)))
## create arbitrary vector of thresholds == 0
sample.th <- rep(0, nVariables)
attr(sample.th, "th.idx") <- 1:nVariables
names(sample.th) <- names(attr(sample.th, "th.idx")) <- paste0(names(KM.ED), " | t1")
## fit model
fit.ED <- sem(mod.ed, sample.cov=KM.ED, sample.nobs = n.ED,
sample.th = sample.th, sample.means = rep(0, nVariables),
estimator="dwls", WLS.V=WLS.V, NACOV = NACOV)You should see estimated thresholds == 0 with nonzero SEs, but you can simply ignore those. I have performed this trick with polychorics from a model estimated in lavaan with DWLS, which I then "forgot" in order to mimic your situation (unknown variances). Fitting the model only to the estimated polychorics and fake thresholds (as above), I obtained identical model results regarding fit and all other parameter estimates.
Granted, this only gets you to the point of using DWLS, but perhaps that is satisfactory. I think you could do a similar hack for full WLS, but that would require more finesse. You would have to interleave fake variances into the NACOV (not a simple block of fake thresholds at the upper left) to provide the order lavaan expects.
Terrence D. Jorgensen
Assistant Professor, Methods and Statistics
Research Institute for Child Development and Education, the University of Amsterdam
Reply all
Reply to author
Forward
0 new messages