Dentatorubrothalamic tracts
27 views
Skip to first unread message
Olivier Roy
Sep 6, 2022, 2:42:25 PM9/6/22
to DSI Studio
Hello Frank,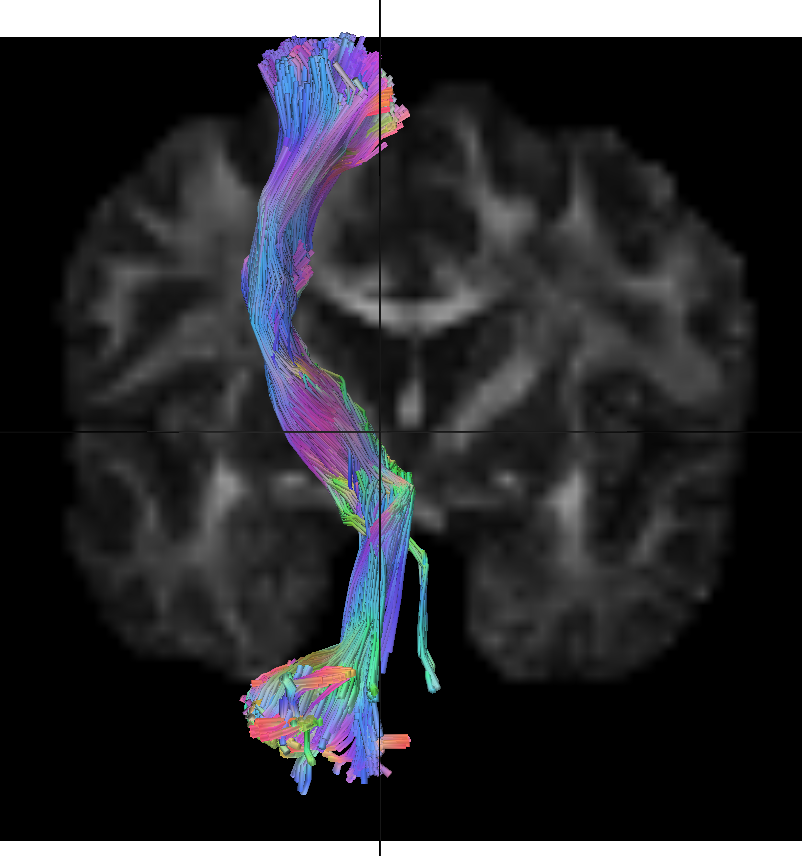
I'm doing tractography of the dentatorubrothalamic on a single subject's data. I'm having a strange issue where when I do autotracking of the right and left tracts, each single tract runs along an ipsilateral path (90% of the fibers). Normally, it should be the other way around as the majority of the fibers from this tract decussate in the red nucleus. Is it possible that this autotrack reconstruction tracks the the dentato-rubro part correctly but then switches to the ipsilateral rubro-thalamo-cortical part afterwards? When displaying both sides, the overall results seem fine.
I've sent the src and fib files to the Dropbox (lesion is known).
Here is also a quick screenshot of what I mean
Thanks,
Olivier

Fang-Cheng Yeh
Sep 6, 2022, 3:33:20 PM9/6/22
to dsi-s...@googlegroups.com
Hi Olivier,
Thank you for reporting this issue. DSI Studio applied a left/right mask to ensure no accidental crossing for any pathway having a label ending with _L or _R
This caused a problem for DRTT. I updated DSI Studio by relabeling it as _LR and _RL so that the left-right mask is disabled.
Please download DSI Studio and see if it works.
Thanks again for reporting the problem.
Frank
--
You received this message because you are subscribed to the Google Groups "DSI Studio" group.
To unsubscribe from this group and stop receiving emails from it, send an email to dsi-studio+...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/9a523c65-de77-4e10-9336-ea6adaf9d065n%40googlegroups.com.
Fang-Cheng Yeh
Sep 6, 2022, 3:33:56 PM9/6/22
to dsi-s...@googlegroups.com
The release should be ready in a few minutes. Status: https://github.com/frankyeh/DSI-Studio/actions/runs/3002662559
Olivier Roy
Sep 7, 2022, 4:24:01 AM9/7/22
to DSI Studio
Hi Frank,
Thanks for the update. I indeed see the updated _LR and _RL labels. However, the resulting reconstructed tracts look similar with few fibers crossing the midline.
Any idea how to resolve the issue?
Thank you very much,
Olivier
Fang-Cheng Yeh
Sep 7, 2022, 9:36:03 AM9/7/22
to dsi-s...@googlegroups.com
You may add ROA or ROI to limit the results and improve the accuracy.
Hope it helps,
Frank
To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/445a126a-ac6b-4c1d-b2f8-d28b9b9332fan%40googlegroups.com.
Olivier Roy
Sep 7, 2022, 9:41:21 AM9/7/22
to DSI Studio
I get your point. However, the reconstruction appears accurate in the sense of the path the tract takes. If it helps, while the tract is being created, I see tracts that arise from the contralateral dentate nucleus and take the overal correct path but they get deleted at the profit of the ipsilateral ones at the end of the process. Could this be the problem? Shouldn't the ipsilateral fibers be deleted instead of the contralateral ones?
Thanks!
Olivier Roy
Sep 12, 2022, 5:09:37 AM9/12/22
to DSI Studio
Hi Frank,
Quick update. After adding ROIs and ROAs to the reconstruction. The vast majority of fibers still do not cross the midline. I'm not sure what is wrong. Do you have an idea? I've sent the data files at the time of my first message posting. Could you take a look?
Thanks!
Olivier
Fang-Cheng Yeh
Sep 12, 2022, 9:56:19 AM9/12/22
to dsi-s...@googlegroups.com
I also got similar results, and it seems that the non-crossing part also includes some reticulospinal tracts and appears much larger. (I don't have a better solution so far).
Frank
To view this discussion on the web visit https://groups.google.com/d/msgid/dsi-studio/0b24322e-4ffa-451c-b441-a18c52a78c97n%40googlegroups.com.
Reply all
Reply to author
Forward
0 new messages
