BioCoder : a high-level programming language for expressing biology protocols
Vaishnavi. A
I am one of the developers of the BioCoder language. BioCoder is
essentially a C library with various functionalities included,
pertaining primarily to the field of Molecular Biology. We envision
the replacement of natural language description of protocols in
published papers with code written in BioCoder, thus making the
protocol standardized and amenable to automation and re-usability.We
are in the process of working out the release of BioCoder.
It is a widely accepted fact that most descriptions of protocols in
published papers are ambiguous and incomplete and we are unsure why
that is the case. We have come up with a survey to try and understand
the problem. It would be great if you could take this short 10-
question survey which tries to zero in on the problem. Results will be
made available on DIYbio when enough data points have been collected.
Please click on the link below to take the survey:
http://www.surveymonkey.com/s/SYJM23Q
For more information about the language:
http://research.microsoft.com/en-us/um/people/thies/thies-iwbda09.pdf
To know what BioCoder code looks like, you can check out the following
examples from OpenWetWare:
http://www.google.co.in/search?q=BioCoder+site%3Aopenwetware.org&ie=utf-8&oe=utf-8&aq=t&client=firefox-a&rlz=1R1WZPB_en-GB___IN356
Owen
English, and it is unambiguous as it is. Encoding your procedure into
C and then decoding it doesn't make much sense to me. C is a tool to
convert higher level ideas into machine instructions. Humans can
understand these high level concepts so there really is no need to do
this.
Bryan Bishop
> The universal language for communicating scientific protocols is
> English, and it is unambiguous as it is. Encoding your procedure into
> C and then decoding it doesn't make much sense to me. C is a tool to
> convert higher level ideas into machine instructions. Humans can
> understand these high level concepts so there really is no need to do
> this.
protocols do not seem to be machine independent, and are highly
function-oriented. It would be interesting to see the protocols
applied to a variety of different machines and different interfaces.
This is what has prompted me to not adopt BioStream immediately:
computational instructions that can't be executed on machines might as
well be useless.
- Bryan
http://heybryan.org/
1 512 203 0507
Eugen Leitl
> The universal language for communicating scientific protocols is
> English, and it is unambiguous as it is. Encoding your procedure into
> C and then decoding it doesn't make much sense to me. C is a tool to
> convert higher level ideas into machine instructions. Humans can
> understand these high level concepts so there really is no need to do
> this.
Machines are notoriously bad at parsing high level concepts, notoriously
good at repeating simple protocols.
As an analogy, consider a random JACS paper vs. a recipe for
a microfluidics reactor (from a library of parts) cum analytics
and inputs as well as outputs and control logic optimized for
a particular synthesis described in above paper.
--
Eugen* Leitl <a href="http://leitl.org">leitl</a> http://leitl.org
______________________________________________________________
ICBM: 48.07100, 11.36820 http://www.ativel.com http://postbiota.org
8B29F6BE: 099D 78BA 2FD3 B014 B08A 7779 75B0 2443 8B29 F6BE
Vaishnavi. A
On Dec 10, 8:10 pm, Bryan Bishop <kanz...@gmail.com> wrote:
> I disagree. The main problem I see with BioStream/BioCoder is that the
> protocols do not seem to be machine independent, and are highly
> function-oriented. It would be interesting to see the protocols
> applied to a variety of different machines and different interfaces.
> This is what has prompted me to not adopt BioStream immediately:
> computational instructions that can't be executed on machines might as
> well be useless.
centric and volume-independent. But then, we didn't have a validation
system in place for this version. To decouple language research from
microfluidics research, we resorted to validation by execution of a
protocol by a biologist and for a biologist to think in terms of
fluids/reagents as opposed to containers and volume-independence, as
opposed to fixed-volume reactions is quite an uphill task. And that is
when we decided to make the language more user-friendly. If you were
to map a container to an arbitrary location (say a microfluidic chip
port) and bring in volume independence again, the language would still
be machine independent.
Vaishnavi. A
On Dec 10, 8:12 pm, Eugen Leitl <eu...@leitl.org> wrote:
> On Thu, Dec 10, 2009 at 06:52:11AM -0800, Owen wrote:
> > The universal language for communicating scientific protocols is
> > English, and it is unambiguous as it is. Encoding your procedure into
>
> Unambiguous, to a skilled person.
>
published in a paper. It is really true that in the first pass one can
get all the details necessary to replicate the protocol successfully?
And this is what we are trying to find out. Why are complete and
precise steps required for the execution of a protocol not published
with the results?
Please take this survey and help us find out!
http://www.surveymonkey.com/s/SYJM23Q
Eugen Leitl
> I disagree. Imagine you had to replicate a protocol that has been
> published in a paper. It is really true that in the first pass one can
> get all the details necessary to replicate the protocol successfully?
critical details. This is rare, but does happen).
> And this is what we are trying to find out. Why are complete and
> precise steps required for the execution of a protocol not published
> with the results?
elsewhere? This is indistinguishable from fraud, and could be
career-ending.
Vaishnavi Ananth
How can you publish something which cannot be easily replicated
elsewhere? This is indistinguishable from fraud, and could be
career-ending.
Anybody who has worked in a wet lab would know what I'm talking about. I'm not saying scientists do not tell people what they did to get their results, they just don't tell you how exactly they did it.
Eugen Leitl
> Anybody who has worked in a wet lab would know what I'm talking about. I'm
> not saying scientists do not tell people what they did to get their results,
> they just don't tell you how exactly they did it.
elsewhere, this will eventually come back to bite you in the ass.
Gabriel Kent
Those who get it now, get it... those who do not, will not, until
there is a simple GUI.
I imagine there is some layer akin to a scene graph? I think that is
what will define its ultimate portability.
JonathanCline
> On Thu, Dec 10, 2009 at 06:52:11AM -0800, Owen wrote:
> > The universal language for communicating scientific protocols is
> > English, and it is unambiguous as it is. Encoding your procedure into
>
> Unambiguous, to a skilled person.
example:
"Incubate 12 hours at room temperature or at 4C overnight."
How long is "overnight" ? Is it 12 hours, or 8, or 6? These are the
kind of ambiguities which are problematic from a machine view.
Also, order-of-operations is confusing from a machine standpoint
(perhaps requires human translation into linear sequence) as in this
example, which mixes present and past tense -- and this is a simple
example:
"“Resuspend pelleted bacterial cells in 250 µl Buffer P1 and transfer
to a micro-centrifuge tube. Ensure that RNase A has been added to
Buffer P1. No cell clumps should be visible after resuspension of the
pellet. If LyseBlue reagent has been added to Buffer P1, vigorously
shake the buffer bottle to ensure LyseBlue particles are completely
dissolved. The bacteria should be resuspended completely by vortexing
or pipetting up and down until no cell clumps remain. "
This is a single protocol step. If the above single step from the
protocol is made into a graph tree, there are ascending and descending
steps required in the parse tree, which isn't what computers are good
at parsing simply (C, for example, is a 1-pass compiler with 1 simple
preprocessor step).
> > C and then decoding it doesn't make much sense to me. C is a tool to
> > convert higher level ideas into machine instructions. Humans can
> > understand these high level concepts so there really is no need to do
> > this.
>
> Did you miss the "amenable to automation and re-usability" part?
> Machines are notoriously bad at parsing high level concepts, notoriously
> good at repeating simple protocols.
mechanism for running automation directly; biologists are not
programmers and should not be expected to learn complex programming
languages (basic principle of "ease of use" is: don't force the
customer to learn new skills to use the system). C is complex for a
complete non-programmer. So I agree that BioStream/BioCoder seems
tangential to the effort (although great work at automated
verification of protocols). Check out the BioStream powerpoint
presentation for real examples of how "compiling" a protocol can leave
ambiguities regarding which step should be completed "next". With all
aspects of the problem in mind, to me it seems that a pseudo-English-
language for running the protocols is the best bet: like a version of
Basic, except for protocols. I prototyped & diagrammed this briefly
here: "Don’t Train the Biology Robot: Have the Machine Read the
Protocol and Automate Itself"
http://88proof.com/synthetic_biology/blog/archives/290
(If someone wants to fund me about $5000 then I'll publish in
PLoS ;-)
The idea of creating a "high level language for protocols" (which C is
not; C is very low level, actually) has been around for a while, even
on the bionet forums going back to early 90's. It hasn't been
realized yet though. In contrast to C, consider the many computer
languages developed over the years which have been specifically
developed to allow ease of use for complex tasks (there are many).
Some variations of BASIC come to mind, although some versions of BASIC
can be quite complex and/or tricky to write instructions in. Logo
also comes to mind, which was created specifically for very ease of
use to teach to children (non-programmers) while providing robotic/
graphical control and was quite successful in its niche. The big
problem of English-language-parsing can be minimized because bio-
protocols use a minimum grammar, that's the key.
Now for the bad news: Suppose Microsoft Research wants to do this
project in order to create a new protocol language for Microsoft,
which they then coerce Current Protocols in Molecular Biology into
using, and everyone else is then forced to obtain a license and/or
expensive software in order to create protocols or even follow
existing protocols. An entire language of biology becomes proprietary
and inaccessible to anyone without the funds or industry clout to
leverage a license; large, and even small, industry projects become
difficult due to continual need to avoid getting stepped on by the
1,000-pound gorilla of Microsoft. And the proprietary and
monopolistic owner, Microsoft, then limits what can, and can not, be
done by the protocol "reader".. such as, sharing protocols or running
protocols on non-Microsoft-approved equipment. That's a bad world,
isn't it. Worse than, and similar to, C# and .NET today, and
Microsoft Windows since version 1.0.
## Jonathan Cline
## jcl...@ieee.org
## Mobile: +1-805-617-0223
########################
Vaishnavi Ananth
I applaud this effort Vaishnavi.
Those who get it now, get it... those who do not, will not, until
there is a simple GUI.
I imagine there is some layer akin to a scene graph? I think that is
what will define its ultimate portability.
Vaishnavi Ananth
Now for the bad news: Suppose Microsoft Research wants to do this
project in order to create a new protocol language for Microsoft,
which they then coerce Current Protocols in Molecular Biology into
using, and everyone else is then forced to obtain a license and/or
expensive software in order to create protocols or even follow
existing protocols. An entire language of biology becomes proprietary
and inaccessible to anyone without the funds or industry clout to
leverage a license; large, and even small, industry projects become
difficult due to continual need to avoid getting stepped on by the
1,000-pound gorilla of Microsoft. And the proprietary and
monopolistic owner, Microsoft, then limits what can, and can not, be
done by the protocol "reader".. such as, sharing protocols or running
protocols on non-Microsoft-approved equipment. That's a bad world,
isn't it. Worse than, and similar to, C# and .NET today, and
Microsoft Windows since version 1.0.
I'd like to make it very clear at this point that BioCoder is not going to be a licensed product. It will be available for free download by the end of December / beginning of January. To put it simply, Microsoft is not going to be "selling" BioCoder.
Doug Treadwell
A good example in the previous discussion was the use of "overnight" to describe a duration. If the ambiguity checker saw this word, it might bring the author to that part of the page (like Word moves the screen to where the grammar error is) and ask them to replace it with a specific time. Like the grammar check it might offer suggestions. "Did you mean: 12 hours; 8 hours; 6 hours?" Any time a lab instrument or reagent is mentioned in the procedures that was not listed in a "materials/equipment" section, it would be noticed and flagged.
Because of the specific contexts, there might be a specific type of grammar the checker could look for. For example, if a sentence read "Resuspend pellet in 250mL solution", the program, which would be smarter than a typical grammar check, would go through a process of determining whether "pellet" and "solution" were clearly identifiable from the preceding text. Also, it might notice that "resuspend in solution" doesn't mention WHERE to resuspend it. So it might say there is a grammar error because insufficient equipment is specified. There might be a procedural grammar database that has a set of equipment requirements for each type of procedure.
All of the above would be hidden from the user, just like Word hides the implementation of the spelling and grammar check from its user. A report author would simply click "ambiguity check" and be shown where their procedure explanation could be improved.
Oh, and if anyone uses this idea and I've added anything to your concept of some product, please give me some credit. I don't want to horde ideas because it delays progress, but having one or two good ideas implemented would be good for my career while the majority of credit would obviously still go to whoever actually implemented them. I'd do the same for you.
- Doug
--
You received this message because you are subscribed to the Google Groups "DIYbio" group.
To post to this group, send email to diy...@googlegroups.com.
To unsubscribe from this group, send email to diybio+un...@googlegroups.com.
For more options, visit this group at http://groups.google.com/group/diybio?hl=en.
Jay Woods
> Perhaps there is another way to provide clarity to descriptions of
> lab procedures. All programming languages that I'm aware of have
> compilers that check for syntax errors. XHTML, which is just a
> lowly markup language, has a free validator at
> http://validator.w3.org/ that checks an XHTML page for usage errors.
> Most word processing programs can check for spelling and grammar
> errors. One way to improve clarity while requiring least effort on
> the part of authors might be to provide a context specific tool for
> improving clarity. It might be called an "ambiguity checker" rather
> than a spell checker.
>
akonadi using the NEPOMUK framework via Soprano from RDF stored in
MySQL. What I haven't been able to figure out is how to fruitfully see
what the triples are that are being stored. The search interface is too
far from what is being proposed here. To me what you are proposing would
be an interface that presents the user with an action list to either
accept the triple (and into the future) as it stands or by presenting
the source of the triple allow the source to be editted into something
where the triples now added to the list are acceptable. It would be
handy if the triples (and source) developed for other protocols could be
used as defaults for the current triple. The KDEPIM includes a journal
feature (as part of Korganizer). The journal could be the place where
all this cross-checking happens. I am currently working with the
journal feature to see if it handles gracefully what I currently handle
with my personal software for notes and plans via tasks, projects, and
protocols.
http://en.wikipedia.org/wiki/NEPOMUK_%28framework%29
http://en.wikipedia.org/wiki/Soprano_%28KDE%29
http://en.wikipedia.org/wiki/Resource_Description_Framework
See Example 3 for the use of XML via RDF tagging to implement the
triples equivalent to the source in English.
http://en.wikipedia.org/wiki/Kdepim - This has a section on Korganizer.
BenjaminPetrin
On Dec 8, 7:26 am, "Vaishnavi. A" <vaishana...@gmail.com> wrote:
> Hi all,
> I am one of the developers of the BioCoder language. BioCoder is
> essentially a C library with various functionalities included,
> pertaining primarily to the field of Molecular Biology.
code I found I'm wondering why the developers chose to implement this
as a C library. Why an imperative language? What advantage does this
offer to simply using a markup language (like XML) and having the
language be the schema (a DTD)?
In either case, as a computer science undergrad with an interest in
biology I look forward to seeing more details on this project.
Vaishnavi Ananth
Vaishnavi Ananth
http://groups.google.com/group/diybio/browse_thread/thread/83ea38b2aef98d8d/b2ea91af6bcd44ca?lnk=raot
BTP Batch2
Nathan McCorkle
I took a look at the BioCoder 1.0 code and got it to compile with make
and gcc on Ubuntu Linux:
https://github.com/nmz787/BioCoder
just make sure you have ncurses-dev installed, something like this:
sudo apt-get install libncurses5-dev libncursesw5-dev
then in the code clone, just run:
make
and the binaries should end up in the Sample_protocol sub-dirs.
Best,
-Nathan
> DIYbio group. To post to this group, send email to diy...@googlegroups.com.
> To unsubscribe from this group, send email to
> diybio+un...@googlegroups.com. For more options, visit this group at
> Learn more at www.diybio.org
> ---
> "DIYbio" group.
> email to diybio+un...@googlegroups.com.
> To view this discussion on the web visit
> https://groups.google.com/d/msgid/diybio/a4271e05-38e7-4715-83ca-a4ffdd2a3ee2%40googlegroups.com.
> For more options, visit https://groups.google.com/d/optout.
--
-Nathan
BTP Batch2
Thomas Meany
Nathan McCorkle
I'm not on the BioCoder team... and only took the ZIP file download of
BioCoder 1.0 and uploaded it to github, then made the changes to get
it to compile on Ubuntu.
You can find the link you referenced here on The Internet Archive:
https://web.archive.org/web/20161110125136/http://research.microsoft.com/en-us/um/india/projects/biocoder/
But I see there it says this same documentation is included in the code repo.
Also, I was the one who created the readme.md file... and only
copy-pasted the journal article title-page text there (along with some
minor formatting).
Please feel free to send me any updates and I can integrate them, or
if you'd like you can simply fork my repo and then I can pull your
changes once you've made any.
Best,
-Nathan
Brian Degger
--
-- You received this message because you are subscribed to the Google Groups DIYbio group. To post to this group, send email to diy...@googlegroups.com. To unsubscribe from this group, send email to diybio+un...@googlegroups.com. For more options, visit this group at https://groups.google.com/d/forum/diybio?hl=en
Learn more at www.diybio.org
---
You received this message because you are subscribed to the Google Groups "DIYbio" group.
To unsubscribe from this group and stop receiving emails from it, send an email to diybio+un...@googlegroups.com.
To post to this group, send email to diy...@googlegroups.com.
Visit this group at https://groups.google.com/group/diybio.
To view this discussion on the web visit https://groups.google.com/d/msgid/diybio/CA%2B82U9K9TP2SGA2Fcp_Rfx68HvQH3uQyqQ5GewHhcoOtsoy9Pw%40mail.gmail.com.
For more options, visit https://groups.google.com/d/optout.
Brian Degger
AdrianMolecule
S James Parsons Jr
--
-- You received this message because you are subscribed to the Google Groups DIYbio group. To post to this group, send email to diy...@googlegroups.com. To unsubscribe from this group, send email to diybio+un...@googlegroups.com. For more options, visit this group at https://groups.google.com/d/forum/diybio?hl=en
Learn more at www.diybio.org
---
You received this message because you are subscribed to the Google Groups "DIYbio" group.
To unsubscribe from this group and stop receiving emails from it, send an email to diybio+un...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/diybio/e0e5dc7d-b0cb-4601-9b12-7909b1061f62%40googlegroups.com.
AdrianMolecule
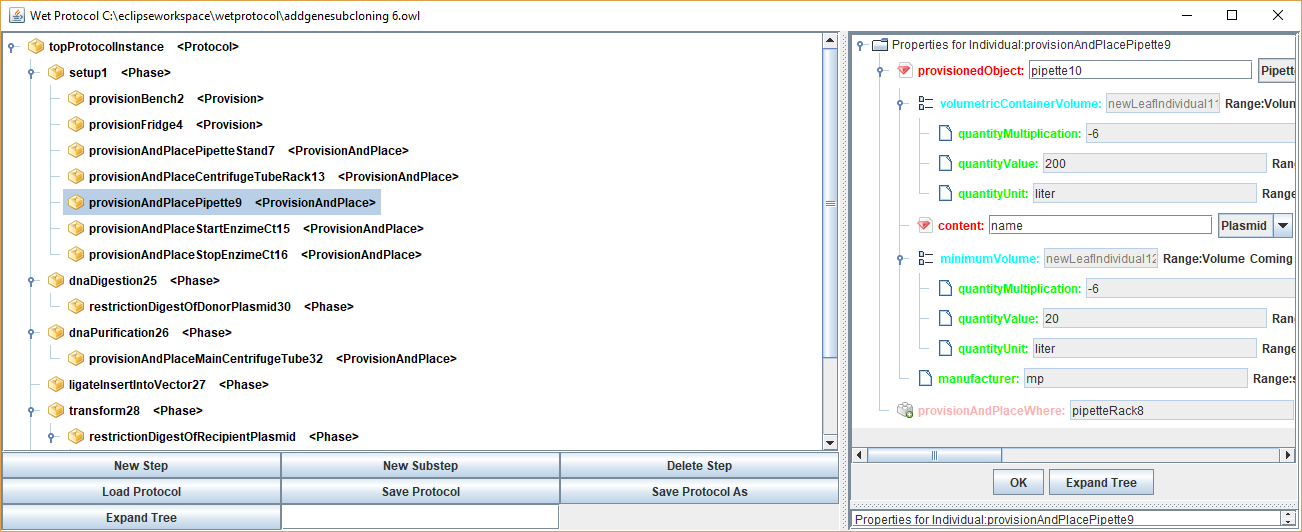
Hi James,
Basically I wanted something that can be as granular that can be used for a simulator or an automated lab directly or could be used for a high level paper.
It's using the Ontology idea (describes the structure in an XML) where the operations (steps) can be added to a global repository edited with an open source editor called
so they can be extended yet standardized. By using a tree one can collapse lower level nodes into more high level protocol.-- You received this message because you are subscribed to the Google Groups DIYbio group. To post to this group, send email to diy...@googlegroups.com. To unsubscribe from this group, send email to diy...@googlegroups.com. For more options, visit this group at https://groups.google.com/d/forum/diybio?hl=en
Learn more at www.diybio.org
---
You received this message because you are subscribed to the Google Groups "DIYbio" group.
To unsubscribe from this group and stop receiving emails from it, send an email to diy...@googlegroups.com.
S James Parsons Jr
On Oct 15, 2019, at 11:14 AM, AdrianMolecule <buzz...@gmail.com> wrote:
Hi James,
Basically I wanted something that can be as granular that can be used for a simulator or an automated lab directly or could be used for a high level paper.
It's using the Ontology idea (describes the structure in an XML) where the operations (steps) can be added to a global repository edited with an open source editor called
so they can be extended yet standardized. By using a tree one can collapse lower level nodes into more high level protocol.
I am considering uploading it on my bio club. URL is specyal.com
Here is a test screenshot.
--
-- You received this message because you are subscribed to the Google Groups DIYbio group. To post to this group, send email to diy...@googlegroups.com. To unsubscribe from this group, send email to diybio+un...@googlegroups.com. For more options, visit this group at https://groups.google.com/d/forum/diybio?hl=en
Learn more at www.diybio.org
---
You received this message because you are subscribed to the Google Groups "DIYbio" group.
To unsubscribe from this group and stop receiving emails from it, send an email to diybio+un...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/diybio/65fa1e09-ecce-4f81-ab4a-eddec59846f3%40googlegroups.com.
<wetprotocol.png>
Nathan McCorkle
Hi James,
Basically I wanted something that can be as granular that can be used for a simulator or an automated lab directly or could be used for a high level paper.
It's using the Ontology idea (describes the structure in an XML)
where the operations (steps) can be added to a global repository
I am considering uploading it