Using year as covariate for anual density estimations
Pamela Narváez
Hello,
I apologize if something like this has been asked before.
We’re working on calculating the densities of a few lemur species over a 5 year period (2015-2019). We are interested on how the densities of these species have changed with changes in forest fragment size, and ideally, we would like to estimate the density of each species per year. We have 37 line transects (length 500m), and we walked them between 6 and 30 times every year.
Towards the last 2 years of the 5-year period, we have been getting fewer and fewer observations (e.g., from 53 in 2015 to 12 in 2019 for one species), so we’re having troubles with some of the results. From what I read in someone else’s email chain, Dr. Buckland mentioned that it’s ok not to have >60 or even >80 sightings in line-transect surveys if you get good fits and the detection function makes sense. However, the observations we have might be a bit too low to continue the analysis (I tried it and it doesn’t look great).
I’ve already tried covariate modeling with rare species following your example (http://examples.distancesampling.org/Distance-spec-covar/species-covariate-distill.html ). However, the density estimates for the more common species appear considerably smaller than what I would get if I run the analysis of those species individually.
I was thinking about combining the 2015-2019 data for each individual species and then using year as a covariate. Do you think this would be a good approach?
Thank you so much for your time!!
Pamela Narváez-TorresUniversity of Calgary
Eric Rexstad
Pamela
Your suggestion about combining data
across years (rather than across species) follows the same
principle. Combining years assumes the detection functions for
all years share the same key function, but the scale parameter
of the key function differs between years. It seems plausible
that detectability for a species shares a key function across
years but that inter-annual differences may cause the scale
parameter to differ between years.
--
You received this message because you are subscribed to the Google Groups "distance-sampling" group.
To unsubscribe from this group and stop receiving emails from it, send an email to distance-sampl...@googlegroups.com.
To view this discussion on the web visit https://groups.google.com/d/msgid/distance-sampling/db391aeb-0709-44bb-b5c7-35314a4abb71n%40googlegroups.com.
-- Eric Rexstad Centre for Ecological and Environmental Modelling University of St Andrews St Andrews is a charity registered in Scotland SC013532
Pamela Narváez
Dear Eric, thank you so much for the reply! I tried doing it for one species, but the densities keep coming up as zero. Here’s the data and code I am using. Any comments would be really helpful!
lem_year<- read.csv(file="Lemur_covyear.csv", sep=",",h=T)
lem_year$year<-as.factor(lem_year$year)
DS_lemY<- ds(data = lem_year,
key="hn",
formula=~year)
summary(DS_lemY)
gof_ds(DS_lemY)
plot.with.covariates(DS_lemY)
ds_cov_lemY <- dht2(ddf=DS_lemY, flatfile=lem_year,
strat_formula = ~year,
stratification = "object")
Thanks again!
Pamela
Eric Rexstad
Pamela
The solution is actually more simple than your approach. For the data you provides, we can treat year as a stratification factor. This simple solution won't work if you have geographical strata as well as yearly surveys. I simply copy your `year` covariate into the `Region.Label` field. This way, there is no need to invoke `dht2`.
I've also fooled around a bit assessing
whether detectability actually changes by year. I've also
introduced a units conversion so that density is reported in
lemurs per sq km.
library(Distance)
lem_year<- read.csv(file="C:/Users/erexs/Documents/My
Distance Projects/Pamela/Lemur_covyear.csv")
conversion <- convert_units("meter", "meter", "square
kilometer")
lem_year$Region.Label <- lem_year$year
DS_lemR<- ds(data = lem_year,
key="hn",
formula=~Region.Label,
convert.units = conversion)
summary(DS_lemR)
gof_ds(DS_lemR)
If you examine the summary() output, you will see year-specific estimates are provided:
Density:
Label Estimate se cv lcl ucl
df
1 2014 10.846999 3.2701748 0.3014820 5.999292 19.611876
50.60852
2 2015 7.193801 2.1466990 0.2984096 3.991637 12.964797
42.98662
3 2016 4.180019 1.2379703 0.2961638 2.336603 7.477762
52.88349
4 2017 4.138706 1.7936978 0.4333959 1.798687 9.522993
49.98898
5 2018 2.755814 1.2932233 0.4692709 1.138530 6.670455
111.57897
6 2019 3.922302 1.8101181 0.4614938 1.623101 9.478433
50.53604
7 Total 5.351963 0.8017811 0.1498107 3.990059 7.178718
213.88215
But the summary also shows the beta parameters associated with the year-specific estimates of the shape parameter of the half-normal key function:
Detection function parametersScale coefficient(s):
estimate se
(Intercept) 2.111800931 0.1330146
Region.Label2015 0.004741692 0.1680775
Region.Label2016 0.327478413 0.2087262
Region.Label2017 0.216059594 0.2441551
Region.Label2018 0.086051672 0.3725973
Region.Label2019 -0.361882282 0.2386520
The standard errors of these betas are of the same size or larger than the estimates themselves. This suggests there is not a strong "year" signal in the shape of the detection functions. Perhaps a model with a pooled detection function would be equally suitable for these data:
DS_lemCons<- ds(data = lem_year,
key="hn", convert.units = conversion)
AIC(DS_lemR, DS_lemCons)
summary(DS_lemCons)
> AIC(DS_lemR, DS_lemCons)
df AIC
DS_lemR 6 960.9689
DS_lemCons 1 959.9827
The AIC comparison implies there is little difference between the models, both of which fit the data.
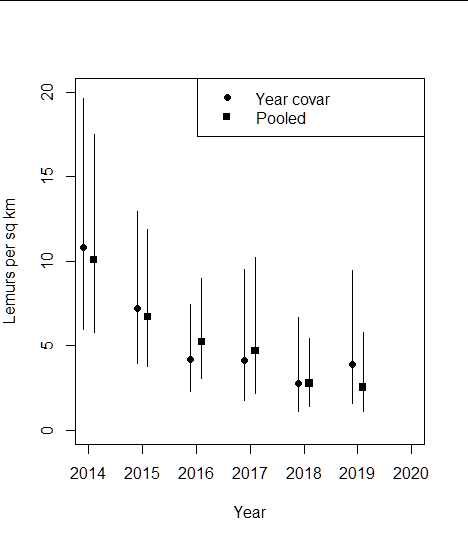
Having a look at the point and interval estimates of the two models bears out the minute difference between the models.
# plottingdens_yrcov <- DS_lemR$dht$individuals$D
dens_yrcov$year <- as.numeric(dens_yrcov$Label) - 0.1
plot(dens_yrcov$year, dens_yrcov$Estimate, ylim=c(0,20), xlim=c(2014,2020), pch=19,
xlab="Year", ylab="Lemurs per sq km")
segments(dens_yrcov$year, dens_yrcov$lcl, dens_yrcov$year, dens_yrcov$ucl)
dens_cons <- DS_lemCons$dht$individuals$D
dens_cons$year <- as.numeric(dens_cons$Label) + 0.1
points(dens_cons$year, dens_cons$Estimate, ylim=c(0,20), pch=15)
segments(dens_cons$year, dens_cons$lcl, dens_cons$year, dens_cons$ucl)
legend("topright", pch=c(19,15), legend=c("Year covar", "Pooled"))
To view this discussion on the web visit https://groups.google.com/d/msgid/distance-sampling/78b3c21f-f750-4d57-9849-dc73f1839472n%40googlegroups.com.
Pamela Narváez
<fdjeehndecciinfo.png>
Hannah Madden
Eric Rexstad
Hannah
Sorry, I can't diagnose your problem from
the information you've provided. Are you willing to send along
the data file (off the list) and I can have a look.
To view this discussion on the web visit https://groups.google.com/d/msgid/distance-sampling/dd916cb4-df10-461d-b4af-58415c1616fan%40googlegroups.com.
Hannah Madden
Pamela Narváez
Eric Rexstad
Pamela
The methods described in Sections 8.5.1
and 8.5.2 of Buckland et al. (2015) are data-hungry. Note the
number of sites in those studies are 446, compared to the 35 you
have. I suspect your habitat measure of interest (forest cover)
does not measurably change between the multiple walks per year;
and as I recall you are unlikely to produce defensible density
estimates on a per-visit basis. This leads me to suspect it
might still be difficult to assess the effect of habitat on
animal density.
Nevertheless, the attached document (for
an upcoming introductory workshop) might be adaptable to your
situation. Before going down this route, you should do lots of
exploratory work to assess whether there might be a signal about
forest cover in your data. The code has not been generalised
hence is likely to require work to conform to your situation.
To view this discussion on the web visit https://groups.google.com/d/msgid/distance-sampling/87335ca1-988c-4fce-8dd6-af83ed933264n%40googlegroups.com.
Pamela Narváez
<countmodel.html>
Jonathan Felis
Eric Rexstad
Jonathan
I'm not sure I can come to grips with all the details of your design and intended estimates, but I'll offer a couple of comments.
Your are correct that two-level stratification is not yet possible with the Distance package. Instead of treating year as a stratum, simply include year as a covariate in the detection function along with other covariates such as region and group size; then employ the strategy you described in your code.
As for using study area-wide estimates of groups size, afraid that cannot be carried out with the Distance package. You could perform the calculation manually using the estimate of mean group size (and standard error) for the entire study area, and combine with stratum-specific estimates of group abundance. However, I would be cautious of this approach. You note there is little evidence of stratum-specific differences in group size, but you note that the data you have from Region 2 is sparse--suggesting you have a poor idea of what Region 2 group size might be. I don't see harm in using region-specific estimates of group size; it reflects the information you have.
In general it does sound like Region 2 is
going to be your burden to bear. Given the survey design with
1/3 the effort allocated to Region 2, you will struggle for
detections.
To view this discussion on the web visit https://groups.google.com/d/msgid/distance-sampling/aa41a04e-8d0e-4256-bb66-a4ba44fdd97cn%40googlegroups.com.
Jonathan Felis
Eric Rexstad
Jonathan
Thanks for your follow-up. You are asking the right questions. For many of your questions, there isn't a "cookbook" answer that says "do this".
The funding constraints resulting in the survey design that you inherited hasn't done you many favours. In the face of sparse data, decisions associated with analysis can have more profound consequences than when a study has a richer pool of data.
I'm comfortable with your suggestion of ignoring the second stratum that is more data poor than the first stratum; that simplifies things a bit. I might also suggest doing the same with sea state, relying upon the pooling robustness property of distance sampling to produce unbiased estimates even when ignoring some factors that might affect detectability.
I don't know what your exploratory analysis might suggest about the effect of group size. Do your organisms congregate in groups of size 1-10 or groups of size 1-10000? If the former, the effect of group size might be small or you could consider treating detection of a group of size 5 as 5 detections of singles all at the same perpendicular distance--thereby diminishing the group size issue.
That might prune off a few issues to make your work more tractable.
Regarding your interest in squeezing further information from the data set, that is an understandable desire, but you've not said anything about the design of the survey to allocate effort among seasons, so you'd need to do exploratory work to see how surveys are temporally distributed within seasons and whether this might be confounded with other factors (crew A surveyed early in the season, while crew B surveyed late in the season; or through time, surveys tended to be conducted earlier (or later) in the season). Given your concern that data are already spread a bit thin, I'm dubious of the chances of a clear picture about seasonal effects upon abundance; but then I'm a pessimist.
For your remaining questions, dealing with
fitting detection functions with possible evasive movement and
varying amounts of heaping, the only general statements I can
make are that you are trying to estimate the shape of the
detection *process*, with vagaries of data quality hindering
that intention. Beyond that, we'd probably be best to go
off-list and report any revelations back to the list.
To view this discussion on the web visit https://groups.google.com/d/msgid/distance-sampling/a8681eb5-a694-41d5-bc7d-356f1dfc1954n%40googlegroups.com.
